Equilibrium unfolding pathway of an H-type RNA pseudoknot which promotes programmed -1 ribosomal frameshifting
- PMID: 10373368
- PMCID: PMC7126474
- DOI: 10.1006/jmbi.1999.2850
Equilibrium unfolding pathway of an H-type RNA pseudoknot which promotes programmed -1 ribosomal frameshifting
Abstract
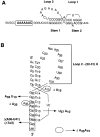
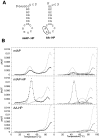
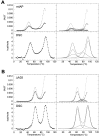
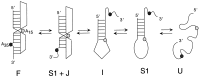
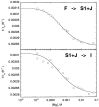
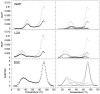
The equilibrium unfolding pathway of a 41-nucleotide frameshifting RNA pseudoknot from the gag-pro junction of mouse intracisternal A-type particles (mIAP), an endogenous retrovirus, has been determined through analysis of dual optical wavelength, equilibrium thermal melting profiles and differential scanning calorimetry. The mIAP pseudoknot is an H-type pseudoknot proposed to have structural features in common with the gag-pro frameshifting pseudoknots from simian retrovirus-1 (SRV-1) and mouse mammary tumor virus (MMTV). In particular, the mIAP pseudoknot is proposed to contain an unpaired adenosine base at the junction of the two helical stems (A15), as well as one in the middle of stem 2 (A35). A mutational analysis of stem 1 hairpins and compensatory base-pair substitutions incorporated into helical stem 2 was used to assign optical melting transitions to molecular unfolding events. The optical melting profile of the wild-type RNA is most simply described by four sequential two-state unfolding transitions. Stem 2 melts first in two closely coupled low-enthalpy transitions at low tmin which the stem 3' to A35, unfolds first, followed by unfolding of the remainder of the helical stem. The third unfolding transition is associated with some type of stacking interactions in the stem 1 hairpin loop not present in the pseudoknot. The fourth transition is assigned to unfolding of stem 1. In all RNAs investigated, DeltaHvH approximately DeltaHcal, suggesting that DeltaCpfor unfolding is small. A35 has the thermodynamic properties expected for an extrahelical, unpaired nucleotide. Deletion of A15 destabilizes the stem 2 unfolding transition in the context of both the wild-type and DeltaA35 mutant RNAs only slightly, by DeltaDeltaG degrees approximately 1 kcal mol-1(at 37 degrees C). The DeltaA15 RNA is considerably more susceptible to thermal denaturation in the presence of moderate urea concentrations than is the wild-type RNA, further evidence of a detectable global destabilization of the molecule. Interestingly, substitution of the nine loop 2 nucleotides with uridine residues induces a more pronounced destabilization of the molecule (DeltaDeltaG degrees approximately 2.0 kcal mol-1), a long-range, non-nearest neighbor effect. These findings provide the thermodynamic basis with which to further refine the relationship between efficient ribosomal frameshifting and pseudoknot structure and stability.
Copyright 1998 Academic Press.
Figures



Similar articles
-
Contribution of the intercalated adenosine at the helical junction to the stability of the gag-pro frameshifting pseudoknot from mouse mammary tumor virus.RNA. 2000 Mar;6(3):409-21. doi: 10.1017/s1355838200992057. RNA. 2000. PMID: 10744025 Free PMC article.
-
Equilibrium unfolding (folding) pathway of a model H-type pseudoknotted RNA: the role of magnesium ions in stability.Biochemistry. 1998 Nov 17;37(46):16116-29. doi: 10.1021/bi981726z. Biochemistry. 1998. PMID: 9819204
-
Mutational analysis of the RNA pseudoknot involved in efficient ribosomal frameshifting in simian retrovirus-1.Nucleic Acids Res. 1998 Mar 15;26(6):1369-72. doi: 10.1093/nar/26.6.1369. Nucleic Acids Res. 1998. PMID: 9490779 Free PMC article.
-
Structure, stability and function of RNA pseudoknots involved in stimulating ribosomal frameshifting.J Mol Biol. 2000 Apr 28;298(2):167-85. doi: 10.1006/jmbi.2000.3668. J Mol Biol. 2000. PMID: 10764589 Free PMC article. Review.
-
Revealing -1 programmed ribosomal frameshifting mechanisms by single-molecule techniques and computational methods.Comput Math Methods Med. 2012;2012:569870. doi: 10.1155/2012/569870. Epub 2012 Apr 1. Comput Math Methods Med. 2012. PMID: 22545064 Free PMC article. Review.
Cited by
-
ProbKnot: fast prediction of RNA secondary structure including pseudoknots.RNA. 2010 Oct;16(10):1870-80. doi: 10.1261/rna.2125310. Epub 2010 Aug 10. RNA. 2010. PMID: 20699301 Free PMC article.
-
Pairwise coupling analysis of helical junction hydrogen bonding interactions in luteoviral RNA pseudoknots.Biochemistry. 2006 Sep 19;45(37):11162-71. doi: 10.1021/bi060430n. Biochemistry. 2006. PMID: 16964977 Free PMC article.
-
Frameshifting RNA pseudoknots: structure and mechanism.Virus Res. 2009 Feb;139(2):193-208. doi: 10.1016/j.virusres.2008.06.008. Epub 2008 Jul 25. Virus Res. 2009. PMID: 18621088 Free PMC article. Review.
-
Single-molecule chemical denaturation of riboswitches.Nucleic Acids Res. 2013 Apr;41(7):4253-65. doi: 10.1093/nar/gkt128. Epub 2013 Feb 27. Nucleic Acids Res. 2013. PMID: 23446276 Free PMC article.
-
Predicting RNA pseudoknot folding thermodynamics.Nucleic Acids Res. 2006 May 18;34(9):2634-52. doi: 10.1093/nar/gkl346. Print 2006. Nucleic Acids Res. 2006. PMID: 16709732 Free PMC article.
References
-
- Banerjee A.R., Jaeger J.A., Turner D.H. Thermal unfolding of a group I ribozyme: the low-temperature transition is primarily disruption of tertiary structure. Biochemistry. 1993;32:153–163. - PubMed
-
- Brierley I. Ribosomal frameshifting on viral RNAs. J. Gen. Virol. 1995;76:1885–1892. - PubMed
-
- Chen X., Chamorro M., Lee S.I., Shen L.X., Hines J.V., Tinoco I., Jr, Varmus H.E. Structural and functional studies of retroviral RNA pseudoknots involved in ribosomal frameshifting: nucleotides at the junction of the two stems are important for efficient ribosomal frameshifting. EMBO J. 1995;14:842–852. - PMC - PubMed
-
- Chen X., Kang H., Shen L.X., Chamorro M., Varmus H.E., Tinoco I., Jr A characteristic bent conformation of RNA pseudoknots promotes −1 frameshifting during translation of retroviral RNA. J. Mol. Biol. 1996;260:479–483. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

