Highly divergent lentiviral Tat proteins activate viral gene expression by a common mechanism
- PMID: 10373508
- PMCID: PMC84257
- DOI: 10.1128/MCB.19.7.4592
Highly divergent lentiviral Tat proteins activate viral gene expression by a common mechanism
Abstract
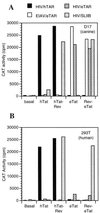
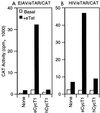
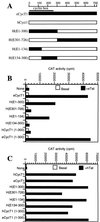
The human immunodeficiency virus type 1 (HIV-1) Tat protein (hTat) activates transcription initiated at the viral long terminal repeat (LTR) promoter by a unique mechanism requiring recruitment of the human cyclin T1 (hCycT1) cofactor to the viral TAR RNA target element. While activation of equine infectious anemia virus (EIAV) gene expression by the EIAV Tat (eTat) protein appears similar in that the target element is a promoter proximal RNA, eTat shows little sequence homology to hTat, does not activate the HIV-1 LTR, and is not active in human cells that effectively support hTat function. To address whether eTat and hTat utilize similar or distinct mechanisms of action, we have cloned the equine homolog of hCycT1 (eCycT1) and examined whether it is required to mediate eTat function. Here, we report that expression of eCycT1 in human cells fully rescues eTat function and that eCycT1 and eTat form a protein complex that specifically binds to the EIAV, but not the HIV-1, TAR element. While hCycT1 is also shown to interact with eTat, the lack of eTat function in human cells is explained by the failure of the resultant protein complex to bind to EIAV TAR. Critical sequences in eCycT1 required to support eTat function are located very close to the amino terminus, i.e., distal to the HIV-1 Tat-TAR interaction motif previously identified in the hCycT1 protein. Together, these data provide a molecular explanation for the species tropism displayed by eTat and demonstrate that highly divergent lentiviral Tat proteins activate transcription from their cognate LTR promoters by essentially identical mechanisms.
Figures

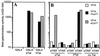

Similar articles
-
An in vitro transcription system that recapitulates equine infectious anemia virus tat-mediated inhibition of human immunodeficiency virus type 1 Tat activity demonstrates a role for positive transcription elongation factor b and associated proteins in the mechanism of Tat activation.Virology. 2000 Sep 1;274(2):356-66. doi: 10.1006/viro.2000.0480. Virology. 2000. PMID: 10964778
-
Interactions between equine cyclin T1, Tat, and TAR are disrupted by a leucine-to-valine substitution found in human cyclin T1.J Virol. 2000 Jan;74(2):892-8. doi: 10.1128/jvi.74.2.892-898.2000. J Virol. 2000. PMID: 10623752 Free PMC article.
-
Canine cyclin T1 rescues equine infectious anemia virus tat trans-activation in human cells.Virology. 2000 Mar 1;268(1):7-11. doi: 10.1006/viro.1999.0141. Virology. 2000. PMID: 10683321
-
Multiple modes of transcriptional regulation by the HIV-1 Tat transactivator.IUBMB Life. 2001 Mar;51(3):175-81. doi: 10.1080/152165401753544241. IUBMB Life. 2001. PMID: 11547919 Review.
-
Regulatory functions of Cdk9 and of cyclin T1 in HIV tat transactivation pathway gene expression.J Cell Biochem. 1999 Dec 1;75(3):357-68. J Cell Biochem. 1999. PMID: 10536359 Review.
Cited by
-
Development of an equine-tropic replication-competent lentivirus assay for equine infectious anemia virus-based lentiviral vectors.Hum Gene Ther Methods. 2012 Oct;23(5):309-23. doi: 10.1089/hgtb.2012.102. Epub 2012 Nov 2. Hum Gene Ther Methods. 2012. PMID: 23121195 Free PMC article.
-
P-TEFb as A Promising Therapeutic Target.Molecules. 2020 Feb 14;25(4):838. doi: 10.3390/molecules25040838. Molecules. 2020. PMID: 32075058 Free PMC article. Review.
-
P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II.Mol Cell Biol. 2000 Apr;20(8):2629-34. doi: 10.1128/MCB.20.8.2629-2634.2000. Mol Cell Biol. 2000. PMID: 10733565 Free PMC article. Review. No abstract available.
-
Mice transgenic for equine cyclin T1 and ELR1 are susceptible to equine infectious anemia virus infection.Retrovirology. 2015 Apr 28;12:36. doi: 10.1186/s12977-015-0163-7. Retrovirology. 2015. PMID: 25928027 Free PMC article.
-
CDK9 autophosphorylation regulates high-affinity binding of the human immunodeficiency virus type 1 tat-P-TEFb complex to TAR RNA.Mol Cell Biol. 2000 Sep;20(18):6958-69. doi: 10.1128/MCB.20.18.6958-6969.2000. Mol Cell Biol. 2000. PMID: 10958691 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
