Major Egr3 isoforms are generated via alternate translation start sites and differ in their abilities to activate transcription
- PMID: 10373520
- PMCID: PMC84269
- DOI: 10.1128/MCB.19.7.4711
Major Egr3 isoforms are generated via alternate translation start sites and differ in their abilities to activate transcription
Abstract
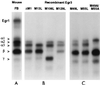
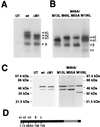
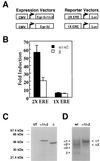
In previous studies, we detected a major, unidentified Egr response element (ERE) binding complex in brain extracts. We now report that this complex contains a truncated isoform of Egr3 generated by use of an alternate translation start site at methionine 106. Furthermore, the ERE binding complex previously thought to contain full-length Egr3 includes several isoforms generated by initiation at other internal methionines. Full-length and truncated (missing residues 1 to 105) Egr3 isoforms differ in the ability to stimulate transcription directed by a tandem repeat of two EREs but not by a single ERE. Taken together, our results indicate that alternative translation start sites are used to generate Egr3 isoforms with distinct transcriptional properties.
Figures


References
-
- Bhat R V, Cole A J, Baraban J M. Chronic cocaine treatment suppresses basal expression of zif268 in rat forebrain: in situ hybridization studies. J Pharmacol Exp Ther. 1992;263:343–349. - PubMed
-
- Bhat R V, Worley P F, Cole A J, Baraban J M. Activation of the zinc finger encoding gene krox-20 in adult rat brain: comparison with zif268. Brain Res Mol Brain Res. 1992;13:263–266. - PubMed
-
- Cole A J, Abu-Shakra S, Saffen D W, Baraban J M, Worley P F. Rapid rise in transcription factor mRNAs in rat brain after electroshock-induced seizures. J Neurochem. 1990;55:1920–1927. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
