Transcriptional cofactor CA150 regulates RNA polymerase II elongation in a TATA-box-dependent manner
- PMID: 10373521
- PMCID: PMC84270
- DOI: 10.1128/MCB.19.7.4719
Transcriptional cofactor CA150 regulates RNA polymerase II elongation in a TATA-box-dependent manner
Abstract
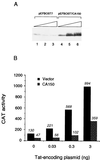
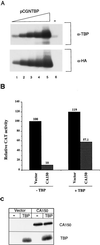
Tat protein strongly activates transcription from the human immunodeficiency virus type 1 (HIV-1) long terminal repeat (LTR) by enhancing the elongation efficiency of RNA polymerase II complexes. Tat-mediated transcriptional activation requires cellular cofactors and specific cis-acting elements within the HIV-1 promoter, among them a functional TATA box. Here, we have investigated the mechanism by which one of these cofactors, termed CA150, regulates HIV-1 transcription in vivo. We present a series of functional assays that demonstrate that the regulation of the HIV-1 LTR by CA150 has the same functional requirements as the activation by Tat. We found that CA150 affects elongation of transcription complexes assembled on the HIV-1 promoter in a TATA-box-dependent manner. We discuss the data in terms of the involvement of CA150 in the regulation of Tat-activated HIV-1 gene expression. In addition, we also provide evidence suggesting a role for CA150 in the regulation of cellular transcriptional processes.
Figures


Similar articles
-
CA150, a nuclear protein associated with the RNA polymerase II holoenzyme, is involved in Tat-activated human immunodeficiency virus type 1 transcription.Mol Cell Biol. 1997 Oct;17(10):6029-39. doi: 10.1128/MCB.17.10.6029. Mol Cell Biol. 1997. PMID: 9315662 Free PMC article.
-
The human immunodeficiency virus type 1 long terminal repeat specifies two different transcription complexes, only one of which is regulated by Tat.J Virol. 1993 Apr;67(4):1752-60. doi: 10.1128/JVI.67.4.1752-1760.1993. J Virol. 1993. PMID: 8445708 Free PMC article.
-
Distinct transcriptional pathways of TAR-dependent and TAR-independent human immunodeficiency virus type-1 transactivation by Tat.Virology. 1997 Aug 18;235(1):48-64. doi: 10.1006/viro.1997.8672. Virology. 1997. PMID: 9300036
-
Tackling Tat.J Mol Biol. 1999 Oct 22;293(2):235-54. doi: 10.1006/jmbi.1999.3060. J Mol Biol. 1999. PMID: 10550206 Review.
-
Multiple modes of transcriptional regulation by the HIV-1 Tat transactivator.IUBMB Life. 2001 Mar;51(3):175-81. doi: 10.1080/152165401753544241. IUBMB Life. 2001. PMID: 11547919 Review.
Cited by
-
The Gln-Ala repeat transcriptional activator CA150 interacts with huntingtin: neuropathologic and genetic evidence for a role in Huntington's disease pathogenesis.Proc Natl Acad Sci U S A. 2001 Feb 13;98(4):1811-6. doi: 10.1073/pnas.98.4.1811. Epub 2001 Jan 30. Proc Natl Acad Sci U S A. 2001. PMID: 11172033 Free PMC article.
-
FF domains of CA150 bind transcription and splicing factors through multiple weak interactions.Mol Cell Biol. 2004 Nov;24(21):9274-85. doi: 10.1128/MCB.24.21.9274-9285.2004. Mol Cell Biol. 2004. PMID: 15485897 Free PMC article.
-
Differential effects of sumoylation on transcription and alternative splicing by transcription elongation regulator 1 (TCERG1).J Biol Chem. 2010 May 14;285(20):15220-15233. doi: 10.1074/jbc.M109.063750. Epub 2010 Mar 9. J Biol Chem. 2010. PMID: 20215116 Free PMC article.
-
Prp40 pre-mRNA processing factor 40 homolog B (PRPF40B) associates with SF1 and U2AF65 and modulates alternative pre-mRNA splicing in vivo.RNA. 2015 Mar;21(3):438-57. doi: 10.1261/rna.047258.114. Epub 2015 Jan 20. RNA. 2015. PMID: 25605964 Free PMC article.
-
RNA polymerase II C-terminal domain: Tethering transcription to transcript and template.Chem Rev. 2013 Nov 13;113(11):8423-55. doi: 10.1021/cr400158h. Epub 2013 Sep 16. Chem Rev. 2013. PMID: 24040939 Free PMC article. Review. No abstract available.
References
-
- Barberis A, Pearlberg J, Simkovich N, Farrell S, Reinagel P, Bamdad C, Sigal G, Ptashne M. Contact with a component of the polymerase II holoenzyme suffices for gene activation. Cell. 1995;81:359–368. - PubMed
-
- Benkirane M, Chun R F, Xiao H, Ogryzko V V, Howard B H, Nakatani Y, Jeang K T. Activation of integrated provirus requires histone acetyltransferase. J Biol Chem. 1998;273:24898–24905. - PubMed
-
- Berger J, Hauber J, Hauber R, Geiger R, Cullen B R. Secreted placental alkaline phosphatase: a powerful new quantitative indicator of gene expression in eukaryotic cells. Gene. 1988;66:1–10. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
