Use of a recombination reporter insert to define meiotic recombination domains on chromosome III of Saccharomyces cerevisiae
- PMID: 10373533
- PMCID: PMC84282
- DOI: 10.1128/MCB.19.7.4832
Use of a recombination reporter insert to define meiotic recombination domains on chromosome III of Saccharomyces cerevisiae
Abstract
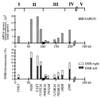
In Saccharomyces cerevisiae, meiotic recombination is initiated by DNA double-strand breaks (DSBs). DSBs usually occur in intergenic regions that display nuclease hypersensitivity in digests of chromatin. DSBs are distributed nonuniformly across chromosomes; on chromosome III, DSBs are concentrated in two "hot" regions, one in each chromosome arm. DSBs occur rarely in regions within about 40 kb of each telomere and in an 80-kb region in the center of the chromosome, just to the right of the centromere. We used recombination reporter inserts containing arg4 mutant alleles to show that the "cold" properties of the central DSB-deficient region are imposed on DNA inserted in the region. Cold region inserts display DSB and recombination frequencies that are substantially less than those seen with similar inserts in flanking hot regions. This occurs without apparent change in chromatin structure, as the same pattern and level of DNase I hypersensitivity is seen in chromatin of hot and cold region inserts. These data are consistent with the suggestion that features of higher-order chromosome structure or chromosome dynamics act in a target sequence-independent manner to control where recombination events initiate during meiosis.
Figures

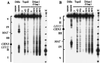



References
-
- Alani E, Padmore R, Kleckner N. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell. 1990;61:419–436. - PubMed
-
- Baker B S, Carpenter A T, Esposito M S, Esposito R E, Sandler L. The genetic control of meiosis. Annu Rev Genet. 1976;10:53–134. - PubMed
-
- Becker D M, Guarente L. High-efficiency transformation of yeast by electroporation. Methods Enzymol. 1991;194:182–187. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
