Homeoproteins CDP and SATB1 interact: potential for tissue-specific regulation
- PMID: 10373541
- PMCID: PMC84297
- DOI: 10.1128/MCB.19.7.4918
Homeoproteins CDP and SATB1 interact: potential for tissue-specific regulation
Abstract
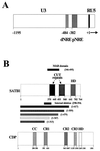
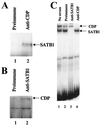
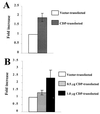
Homeoproteins are known to participate in development and cell type specification. The homeoproteins CCAAT displacement protein (CDP) and special AT-rich sequence binding protein 1 (SATB1) have been shown to bind to nuclear matrix-associated regions and to act as repressors of many cellular genes. Moreover, binding of SATB1 to the mouse mammary tumor virus (MMTV) promoter region dramatically affects the tissue-specific transcription of this retrovirus. Because protein-protein interactions are a common means of regulating homeoprotein function, we tested whether SATB1 and CDP interact in vivo and in vitro. SATB1 interacted with CDP through its DNA-binding domain, as demonstrated by glutathione S-transferase (GST) pull-down assays. GST pull-down assays also showed that CDP associated with SATB1 through three of its four DNA-binding domains (CR1, CR2, and the homeodomain). SATB1-specific antisera, but not preimmune sera, precipitated CDP from nuclear extracts, and CDP-specific antisera precipitated SATB1 from the same extracts. Far-Western blotting detected interaction of SATB1 and CDP in several different tissue extracts. Association of purified SATB1 and CDP in vitro resulted in the inability of each protein to bind to DNA in gel retardation assays. CDP overexpression in cultured T cells led to a loss of detectable SATB1 binding to the MMTV promoter region, as measured by gel shift experiments. CDP overexpression also elevated MMTV long terminal repeat reporter gene activity in transient-transfection assays, a result consistent with neutralization of the SATB1 repressor function in T cells. SATB1 is very abundant in certain tissues, particularly thymus, whereas CDP is relatively ubiquitous, except in certain terminally differentiated cell types. Because of the tissue and cell type distribution of SATB1 and CDP, we propose that the SATB1-to-CDP ratio in different tissues is a novel mechanism for homeoproteins to control gene expression and differentiation in mammals.
Figures
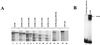
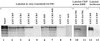

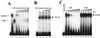
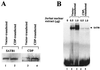

References
-
- Andres V, Nadal-Ginard B, Mahdavi V. Clox, a mammalian homeobox gene related to Drosophila cut, encodes DNA-binding regulatory proteins differentially expressed during development. Development. 1992;116:321–334. - PubMed
-
- Banan M, Rojas I C, Lee W H, King H L, Harriss J V, Kobayashi R, Webb C F, Gottlieb P D. Interaction of the nuclear matrix-associated region (MAR)-binding proteins, SATB1 and CDP/Cux, with a MAR element (L2a) in an upstream regulatory region of the mouse CD8α gene. J Biol Chem. 1997;272:18440–18452. - PubMed
-
- Barberis A, Superti-Furga G, Busslinger M. Mutually exclusive interaction of the CCAAT-binding factor and of a displacement protein with overlapping sequences of a histone gene promoter. Cell. 1987;50:347–359. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
