Utilization of splicing elements and polyadenylation signal elements in the coupling of polyadenylation and last-intron removal
- PMID: 10373547
- PMCID: PMC84315
- DOI: 10.1128/MCB.19.7.4971
Utilization of splicing elements and polyadenylation signal elements in the coupling of polyadenylation and last-intron removal
Abstract
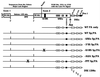
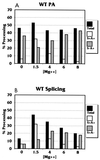
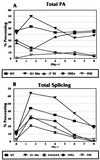
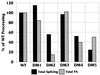
Polyadenylation (PA) is the process by which the 3' ends of most mammalian mRNAs are formed. In nature, PA is highly coordinated, or coupled, with splicing. In mammalian systems, the most compelling mechanistic model for coupling arises from data supporting exon definition (2, 34, 37). We have examined the roles of individual functional components of splicing and PA signals in the coupling process by using an in vitro splicing and PA reaction with a synthetic pre-mRNA substrate containing an adenovirus splicing cassette and the simian virus 40 late PA signal. The effects of individually mutating splicing elements and PA elements in this substrate were determined. We found that mutation of the polypyrimidine tract and the 3' splice site significantly reduced PA efficiency and that mutation of the AAUAAA and the downstream elements of the PA signal decreased splicing efficiency, suggesting that these elements are the most significant for the coupling of splicing and PA. Although mutation of the upstream elements (USEs) of the PA signal dramatically decreased PA, splicing was only modestly affected, suggesting that USEs modestly affect coupling. Mutation of the 5' splice site in the presence of a viable polypyrimidine tract and the 3' splice site had no effect on PA, suggesting no effect of this element on coupling. However, our data also suggest that a site for U1 snRNP binding (e.g., a 5' splice site) within the last exon can negatively effect both PA and splicing; hence, a 5' splice site-like sequence in this position appears to be a modulator of coupling. In addition, we show that the RNA-protein complex formed to define an exon may inhibit processing if the definition of an adjacent exon fails. This finding indicates a mechanism for monitoring the appropriate definition of exons and for allowing only pre-mRNAs with successfully defined exons to be processed.
Figures




References
-
- Berget S M. Exon recognition in vertebrate splicing. J Biol Chem. 1995;270:2411–2414. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
