Defects in components of the proteasome enhance transcriptional silencing at fission yeast centromeres and impair chromosome segregation
- PMID: 10373564
- PMCID: PMC84358
- DOI: 10.1128/MCB.19.7.5155
Defects in components of the proteasome enhance transcriptional silencing at fission yeast centromeres and impair chromosome segregation
Abstract
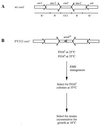
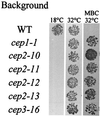
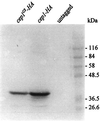
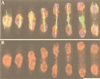
Fission yeast centromeres are transcriptionally silent and form a heterochromatin-like structure essential for normal centromere function; this appears analogous to heterochromatin and position effect variegation in other eukaryotes. Conditional mutations in three genes designated cep (centromere enhancer of position effect) were found to enhance transcriptional silencing within centromeres. Cloning of the cep1(+) and cep2(+) genes by functional complementation revealed that they are identical to the previously described genes pad1(+) and mts2(+), respectively, which both encode subunits of the proteasome 19S cap. Like Mts2 and Mts4, epitope-tagged Cep1/Pad1 localizes to or near the nuclear envelope throughout the cell cycle. The cep mutants display a range of phenotypes depending on the temperature. Silencing within the central domain of centromeres is increased at 36 degrees C. This suggests that the proteasome is involved in regulating silencing and thus centromeric chromatin architecture, possibly by lowering the level of some chromatin-associated protein by ubiquitin-dependent degradation. This is the first report of defective proteasome function affecting heterochromatin-mediated transcriptional silencing. At 36 and 32 degrees C, the cep mutants lose chromosomes at an elevated rate, and at 18 degrees C, the mutants are cryosensitive for growth. Cytological analysis at 18 degrees C revealed a defect in sister chromatid separation while other mitotic events occurred normally, indicating that cep mutations might interfere specifically with the degradation of inhibitor(s) of sister chromatid separation. These observations suggest that 19S subunits confer a level of substrate specificity on the proteasome and raise the possibility of a link between components involved in centromere architecture and sister chromatid cohesion.
Figures


Similar articles
-
Distinct centromere domain structures with separate functions demonstrated in live fission yeast cells.J Cell Sci. 2003 Oct 1;116(Pt 19):4035-42. doi: 10.1242/jcs.00707. Epub 2003 Aug 19. J Cell Sci. 2003. PMID: 12928332
-
Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP-C.Mol Biol Cell. 1995 Jul;6(7):793-807. doi: 10.1091/mbc.6.7.793. Mol Biol Cell. 1995. PMID: 7579695 Free PMC article.
-
Requirement of heterochromatin for cohesion at centromeres.Science. 2001 Dec 21;294(5551):2539-42. doi: 10.1126/science.1064027. Epub 2001 Oct 11. Science. 2001. PMID: 11598266
-
Dissecting Fission Yeast Centromeres via Silencing.In: Mapping Protein/DNA Interactions by Cross-Linking [Internet]. Paris: Institut national de la santé et de la recherche médicale; 2001. In: Mapping Protein/DNA Interactions by Cross-Linking [Internet]. Paris: Institut national de la santé et de la recherche médicale; 2001. PMID: 21413369 Free Books & Documents. Review.
-
The centromere of budding yeast.Bioessays. 1993 Jul;15(7):451-60. doi: 10.1002/bies.950150704. Bioessays. 1993. PMID: 8379948 Review.
Cited by
-
The fission yeast ubiquitin-conjugating enzymes UbcP3, Ubc15, and Rhp6 affect transcriptional silencing of the mating-type region.Eukaryot Cell. 2002 Aug;1(4):613-25. doi: 10.1128/EC.1.4.613-625.2002. Eukaryot Cell. 2002. PMID: 12456009 Free PMC article.
-
Kinetochore and heterochromatin domains of the fission yeast centromere.Chromosome Res. 2004;12(6):521-34. doi: 10.1023/B:CHRO.0000036586.81775.8b. Chromosome Res. 2004. PMID: 15289660 Review.
-
The 19S proteasome is directly involved in the regulation of heterochromatin spreading in fission yeast.J Biol Chem. 2017 Oct 13;292(41):17144-17155. doi: 10.1074/jbc.M117.790824. Epub 2017 Aug 7. J Biol Chem. 2017. PMID: 28784663 Free PMC article.
-
Transcriptional silencing in Saccharomyces cerevisiae and Schizosaccharomyces pombe.Nucleic Acids Res. 2002 Apr 1;30(7):1465-82. doi: 10.1093/nar/30.7.1465. Nucleic Acids Res. 2002. PMID: 11917007 Free PMC article. Review.
-
Chromosomal position effects reveal different cis-acting requirements for rDNA transcription and sex chromosome pairing in Drosophila melanogaster.Genetics. 2000 Jul;155(3):1195-211. doi: 10.1093/genetics/155.3.1195. Genetics. 2000. PMID: 10880481 Free PMC article.
References
-
- Allshire R C, Javerzat J-P, Redhead N J, Cranston G. Position effect variegation at fission yeast centromeres. Cell. 1994;76:157–169. - PubMed
-
- Allshire R C, Nimmo E R, Ekwall K, Javerzat J-P, Cranston G. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 1995;9:218–233. - PubMed
-
- Altshul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Beach D, Piper M, Nurse P. Construction of a Schizosaccharomyces pombe gene bank in a yeast bacterial shuttle vector and its use to isolate genes by complementation. Mol Gen Genet. 1982;187:326–329. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
