An invariant T cell receptor alpha chain defines a novel TAP-independent major histocompatibility complex class Ib-restricted alpha/beta T cell subpopulation in mammals
- PMID: 10377186
- PMCID: PMC2192962
- DOI: 10.1084/jem.189.12.1907
An invariant T cell receptor alpha chain defines a novel TAP-independent major histocompatibility complex class Ib-restricted alpha/beta T cell subpopulation in mammals
Abstract
We describe here a new subset of T cells, found in humans, mice, and cattle. These cells bear a canonical T cell receptor (TCR) alpha chain containing hAV7S2 and AJ33 in humans and the homologous AV19-AJ33 in mice and cattle with a CDR3 of constant length. These T cells are CD4(-)CD8(-) double-negative (DN) T cells in the three species and also CD8alphaalpha in humans. In humans, their frequency was approximately 1/10 in DN, 1/50 in CD8alpha+, and 1/6,000 in CD4(+) lymphocytes, and they display an activated/memory phenotype (CD45RAloCD45RO+). They preferentially use hBV2S1 and hBV13 segments and have an oligoclonal Vbeta repertoire suggesting peripheral expansions. These cells were present in major histocompatibility complex (MHC) class II- and transporter associated with antigen processing (TAP)-deficient humans and mice and also in classical MHC class I- and CD1-deficient mice but were absent from beta2-microglobulin-deficient mice, indicating their probable selection by a nonclassical MHC class Ib molecule distinct from CD1. The conservation between mammalian species, the abundance, and the unique selection pattern suggest an important role for cells using this novel canonical TCR alpha chain.
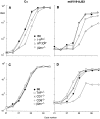
Figures







References
-
- Davis, M.M., and Y.-H. Chien. 1998. T-cell antigen receptors. In Fundamental Immunology. 4th ed. W.E. Paul, editor. Lippincott-Raven Publishers, Philadelphia. 341–366.
-
- Hardy RR, Li YS, Hayakawa K. Distinctive developmental origins and specificities of the CD5+ B-cell subset. Semin Immunol. 1996;8:37–44. - PubMed
-
- Kantor AB, Herzenberg LA. Origin of murine B cell lineages. Annu Rev Immunol. 1993;11:501–538. - PubMed
-
- Haas W, Pereira P, Tonegawa S. Gamma/delta cells. Annu Rev Immunol. 1993;11:637–685. - PubMed
-
- Bendelac A, Rivera MN, Park SH, Roark JH. Mouse CD1-specific NK1 T cells: development, specificity, and function. Annu Rev Immunol. 1997;15:535–562. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

