NFAT5, a constitutively nuclear NFAT protein that does not cooperate with Fos and Jun
- PMID: 10377394
- PMCID: PMC22056
- DOI: 10.1073/pnas.96.13.7214
NFAT5, a constitutively nuclear NFAT protein that does not cooperate with Fos and Jun
Abstract
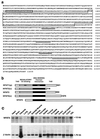
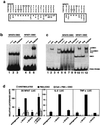
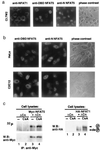
NFAT transcription factors are related to NF-kappaB/Rel proteins and form cooperative complexes with Fos and Jun on DNA. We have identified an NFAT-related protein, NFAT5, which differs from the conventional NFAT proteins NFAT1-4 in its structure, DNA binding, and regulation. NFAT5 contains a NFAT-like Rel homology domain, conserves the DNA contact residues of NFAT1-4, and binds DNA sequences similar to those found in the regulatory regions of well-characterized NFAT-dependent genes. However, it lacks the majority of Fos/Jun contact residues and does not bind cooperatively with Fos and Jun to DNA. Unlike NFAT1-4, whose nuclear import is tightly regulated by calcineurin-mediated dephosphorylation, NFAT5 is a constitutively nuclear phosphoprotein regardless of calcineurin activation. These features suggest that unlike the conventional NFAT proteins, NFAT1-4, which activate gene transcription by integrating inputs from calcium/calcineurin and protein kinase C/mitogen-activated protein kinase signaling pathways, NFAT5 participates in as-yet-unidentified signaling pathways in diverse immune and nonimmune cells.
Figures

References
-
- McCaffrey P G, Luo C, Kerppola T K, Jain J, Badalian T M, Ho A M, Burgeon E, Lane W S, Lambert J N, Curran T, et al. Science. 1993;262:750–754. - PubMed
-
- Northrop J P, Ho S N, Chen L, Thomas D J, Timmerman L A, Nolan G P, Admon A, Crabtree G R. Nature (London) 1994;369:497–502. - PubMed
-
- Park J, Takeuchi A, Sharma S. J Biol Chem. 1996;271:20914–20921. - PubMed
-
- Chuvpilo C, Zimmer M, Kerstan A, Glockner J, Avots A, Escher C, Fischer C, Jankevikcs E, Berberich-Siebelt F, Schmitt E, Serfling E. Immunity. 1999;10:261–269. - PubMed
-
- Hoey T, Sun Y L, Williamson K, Xu X. Immunity. 1995;2:461–472. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

