Identification of human prostaglandin E synthase: a microsomal, glutathione-dependent, inducible enzyme, constituting a potential novel drug target
- PMID: 10377395
- PMCID: PMC22058
- DOI: 10.1073/pnas.96.13.7220
Identification of human prostaglandin E synthase: a microsomal, glutathione-dependent, inducible enzyme, constituting a potential novel drug target
Abstract
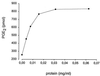
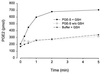
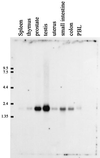
Human prostaglandin (PG) E synthase (EC 5.3.99.3) is a member of a recently recognized protein superfamily consisting of membrane associated proteins involved in eicosanoid and glutathione metabolism (the MAPEG family). Previous designations of the protein are PIG12 and MGST1-L1. PGE synthase was expressed in Escherichia coli, and both cytosolic and membrane fractions were prepared. Western blot analysis specifically detected a 15- to 16-kDa protein in the membrane fraction. Both fractions were incubated with prostaglandin H2 in the presence or absence of reduced glutathione. The membrane but not the cytosolic fraction was found to possess high glutathione-dependent PGE synthase activity (0.25 micromol/min/mg). The human tissue distribution was analyzed by Northern blot analysis. High expression of PGE synthase mRNA was detected in A549 and HeLa cancer cell lines. Intermediate level of expression was demonstrated in placenta, prostate, testis, mammary gland, and bladder whereas low mRNA expression was observed in several other tissues. A549 cells have been used as a model system to study cyclooxygenase-2 induction by IL-1beta. If A549 cells were grown in the presence of IL-1beta, a significant induction of the PGE synthase was observed by Western blot analysis. Also, Western blot analysis specifically detected a 16-kDa protein in sheep seminal vesicles. In summary, we have identified a human membrane bound PGE synthase. The enzyme activity is glutathione-dependent, and the protein expression is induced by the proinflammatory cytokine IL-1beta. PGE synthase is a potential novel target for drug development.
Figures





References
-
- Smith W. Adv Exp Med Biol. 1997;400B:989–1011. - PubMed
-
- Herschman H R. Biochim Biophys Acta. 1996;1299:125–140. - PubMed
-
- Dubois R, Abramson S, Crofford L, Gupta R, Simon L, Van de Putte L, Lipsky P. FASEB J. 1998;12:1063–1073. - PubMed
-
- Smith W L. Am J Physiol. 1992;263:F181–F191. - PubMed
-
- Hamberg M, Samuelsson B. J Biol Chem. 1967;242:5336–5343. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

