The active form of the steroidogenic acute regulatory protein, StAR, appears to be a molten globule
- PMID: 10377400
- PMCID: PMC22068
- DOI: 10.1073/pnas.96.13.7250
The active form of the steroidogenic acute regulatory protein, StAR, appears to be a molten globule
Abstract
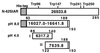
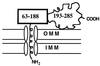
The steroidogenic acute regulatory protein (StAR) increases the movement of cholesterol from the outer to the inner membrane of adrenal and gonadal mitochondria, thus providing the substrate for steroid hormone biosynthesis. Deletion of 62 amino-terminal aa produces a cytoplasmic form of StAR (N-62 StAR) that lacks the mitochondrial leader sequence but retains full activity and appears to act at the outer mitochondrial membrane. At neutral pH the native state of bacterially expressed N-62 StAR protein displays cooperative unfolding under the influence of urea with DeltaGH2O = -4.1 kcal/mol, and it remains correctly folded down to pH 4. Limited proteolysis at different pHs shows that the biologically essential C-terminal region is accessible to solvent, and that the N-terminal domain is compact at pH 8 and partially unfolds below pH 4. Secondary structural analysis of CD curves suggests that the unfolding may coincide with an increase in alpha-helical character at pH 3.5. Fluorescence spectroscopy at pH 3-8 and at 0-6 M urea is consistent with two distinct domains, a compact N-terminal domain containing tryptophans 96 and 147 and a more solvent-accessible C-terminal domain containing tryptophans 241 and 250. These observations suggest that StAR forms a molten globule structure at pH 3.5-4.0. As the mitochondrial proton pump results in an electrochemical gradient, and as StAR must unfold during mitochondrial entry, StAR probably undergoes a similar conformational shift to an extended structure while interacting with the mitochondrial outer membrane, allowing this apparent molten globule form to act as an on/off switch for cholesterol entry into the mitochondria.
Figures

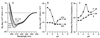

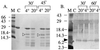
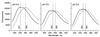
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

