Normal growth and development in the absence of hepatic insulin-like growth factor I
- PMID: 10377413
- PMCID: PMC22084
- DOI: 10.1073/pnas.96.13.7324
Normal growth and development in the absence of hepatic insulin-like growth factor I
Abstract
The somatomedin hypothesis proposed that insulin-like growth factor I (IGF-I) was a hepatically derived circulating mediator of growth hormone and is a crucial factor for postnatal growth and development. To reassess this hypothesis, we have used the Cre/loxP recombination system to delete the igf1 gene exclusively in the liver. igf1 gene deletion in the liver abrogated expression of igf1 mRNA and caused a dramatic reduction in circulating IGF-I levels. However, growth as determined by body weight, body length, and femoral length did not differ from wild-type littermates. Although our model proves that hepatic IGF-I is indeed the major contributor to circulating IGF-I levels in mice it challenges the concept that circulating IGF-I is crucial for normal postnatal growth. Rather, our model provides direct evidence for the importance of the autocrine/paracrine role of IGF-I.
Figures

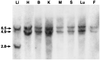
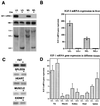
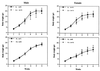
References
-
- Daughaday W H, Rotwein P. Endocr Rev. 1989;10:68–91. - PubMed
-
- Rajaram S, Baylink D J, Mohan S. Endocr Rev. 1997;18:801–831. - PubMed
-
- LeRoith D, Werner H, Beitner-Johnson D, Roberts C T., Jr Endocr Rev. 1995;16:143–163. - PubMed
-
- Liu J P, Baker J, Perkins A S, Robertson E J, Efstratiadis A. Cell. 1993;75:59–72. - PubMed
-
- Baker J, Liu J P, Robertson E J, Efstratiadis A. Cell. 1993;75:73–82. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

