Conformational variants of class II MHC/peptide complexes induced by N- and C-terminal extensions of minimal peptide epitopes
- PMID: 10377434
- PMCID: PMC22105
- DOI: 10.1073/pnas.96.13.7445
Conformational variants of class II MHC/peptide complexes induced by N- and C-terminal extensions of minimal peptide epitopes
Abstract
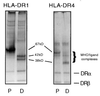
Class II MHC molecules are known to exist in conformational variants. "Floppy" and "compact" forms of murine MHC molecules, for example, are discriminated by their migration behavior on SDS/PAGE and represent empty and ligand-loaded forms. Here we show that formation of distinctly faster-migrating ligand complexes (F-forms) rather than the normal compact (C-) forms of HLA-DR1 or -DR4 results from extensions of minimal peptide epitopes (such as HA306-318 or IC106-120) by approximately 10 amino acids at either the N or the C terminus. Two similar but distinct F-forms (FI and FII) were detected, depending on the site of the extension. Both F-forms were characterized by increased surface hydrophobicity and reduced SDS-stability. Native gel separations and size exclusion chromatography indicated that the F-forms had increased hydrodynamic radii compared with the C-form and an apparent size similar to that of empty MHC molecules. The regions on the ligand overhangs responsible for the effect began at a distance of approximately 5 amino acids on either side of the epitopes, comprised 4-8 amino acids (i.e., a total overhang of 9-14), and did not have a particular sequence preference. The possible functional significance of these forms is discussed.
Figures

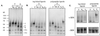


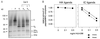
Similar articles
-
Recognition of core and flanking amino acids of MHC class II-bound peptides by the T cell receptor.Eur J Immunol. 2002 Sep;32(9):2510-20. doi: 10.1002/1521-4141(200209)32:9<2510::AID-IMMU2510>3.0.CO;2-Q. Eur J Immunol. 2002. PMID: 12207335
-
A recombinant single-chain human class II MHC molecule (HLA-DR1) as a covalently linked heterotrimer of alpha chain, beta chain, and antigenic peptide, with immunogenicity in vitro and reduced affinity for bacterial superantigens.Eur J Immunol. 1997 Aug;27(8):1933-41. doi: 10.1002/eji.1830270817. Eur J Immunol. 1997. PMID: 9295029
-
Susceptibility to HLA-DM protein is determined by a dynamic conformation of major histocompatibility complex class II molecule bound with peptide.J Biol Chem. 2014 Aug 22;289(34):23449-64. doi: 10.1074/jbc.M114.585539. Epub 2014 Jul 7. J Biol Chem. 2014. PMID: 25002586 Free PMC article.
-
Antigen presentation. Getting peptides into MHC class II molecules.Curr Biol. 1994 Jun 1;4(6):541-3. doi: 10.1016/s0960-9822(00)00119-6. Curr Biol. 1994. PMID: 7922377 Review.
-
Structure of peptides associated with class I and class II MHC molecules.Annu Rev Immunol. 1994;12:181-207. doi: 10.1146/annurev.iy.12.040194.001145. Annu Rev Immunol. 1994. PMID: 7516668 Review.
Cited by
-
Immunogenic peptide discovery in cancer genomes.Curr Opin Genet Dev. 2015 Feb;30:7-16. doi: 10.1016/j.gde.2014.12.003. Epub 2015 Jan 12. Curr Opin Genet Dev. 2015. PMID: 25588790 Free PMC article. Review.
-
Prediction of peptides binding to MHC class I and II alleles by temporal motif mining.BMC Bioinformatics. 2013;14 Suppl 2(Suppl 2):S13. doi: 10.1186/1471-2105-14-S2-S13. Epub 2013 Jan 21. BMC Bioinformatics. 2013. PMID: 23368521 Free PMC article.
-
A pH-sensitive histidine residue as control element for ligand release from HLA-DR molecules.Proc Natl Acad Sci U S A. 2002 Dec 24;99(26):16946-50. doi: 10.1073/pnas.212643999. Epub 2002 Dec 5. Proc Natl Acad Sci U S A. 2002. PMID: 12471156 Free PMC article.
-
Paradoxical intrathymic positive selection in mice with only a covalently presented agonist peptide.Proc Natl Acad Sci U S A. 2001 Jul 31;98(16):9243-8. doi: 10.1073/pnas.161274698. Epub 2001 Jul 24. Proc Natl Acad Sci U S A. 2001. PMID: 11470911 Free PMC article.
-
Structural Insights Into HLA-DM Mediated MHC II Peptide Exchange.Curr Top Biochem Res. 2011;13(2):39-55. Curr Top Biochem Res. 2011. PMID: 25264402 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

