JC virus DNA is present in the mucosa of the human colon and in colorectal cancers
- PMID: 10377441
- PMCID: PMC22112
- DOI: 10.1073/pnas.96.13.7484
JC virus DNA is present in the mucosa of the human colon and in colorectal cancers
Abstract
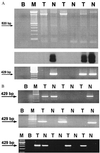
JC virus (JCV) is a polyoma virus that commonly infects humans. We have found T antigen DNA sequences of JCV in the mucosa of normal human colons, colorectal cancers, colorectal cancer xenografts raised in nude mice, and in the human colon cancer cell line SW480. A larger number of viral copies is present in cancer cells than in non-neoplastic colon cells, and sequence microheterogeneity occurs within individual colonic mucosal specimens. The improved yield of detection after treatment with topoisomerase I suggests that the viral DNA is negatively supercoiled in the human tissues. These results indicate that JCV DNA can be found in colonic tissues, which raises the possibility that this virus may play a role in the chromosomal instability observed in colorectal carcinogenesis.
Figures



Comment in
-
Convicting a human tumor virus: guilt by association?Proc Natl Acad Sci U S A. 1999 Jul 6;96(14):7619-21. doi: 10.1073/pnas.96.14.7619. Proc Natl Acad Sci U S A. 1999. PMID: 10393867 Free PMC article. Review. No abstract available.
-
JC virus and human colon carcinoma: an intriguing and inconclusive association.Gastroenterology. 2003 Jan;124(1):268-9; author reply 269-70. doi: 10.1053/gast.2003.50033. Gastroenterology. 2003. PMID: 12512060 No abstract available.
Similar articles
-
Detection of JC virus DNA sequences in colorectal cancers in Japan.Virchows Arch. 2005 Oct;447(4):723-30. doi: 10.1007/s00428-005-0014-3. Epub 2005 Oct 19. Virchows Arch. 2005. PMID: 16021515
-
Human JC polyomavirus in normal colorectal mucosa, hyperplastic polyps, sporadic adenomas, and adenocarcinomas in Portugal.J Med Virol. 2013 Dec;85(12):2119-27. doi: 10.1002/jmv.23705. Epub 2013 Sep 5. J Med Virol. 2013. PMID: 24009184
-
Association of JC virus T-antigen expression with the methylator phenotype in sporadic colorectal cancers.Gastroenterology. 2006 Jun;130(7):1950-61. doi: 10.1053/j.gastro.2006.02.061. Gastroenterology. 2006. PMID: 16762618
-
JC virus and colorectal cancer: a possible trigger in the chromosomal instability pathways.Curr Opin Gastroenterol. 2005 Jan;21(1):85-9. Curr Opin Gastroenterol. 2005. PMID: 15687890 Review.
-
JC virus in the pathogenesis of colorectal cancer, an etiological agent or another component in a multistep process?Virol J. 2010 Feb 18;7:42. doi: 10.1186/1743-422X-7-42. Virol J. 2010. PMID: 20167111 Free PMC article. Review.
Cited by
-
Differential expression of viral agents in lymphoma tissues of patients with ABC diffuse large B-cell lymphoma from high and low endemic infectious disease regions.Oncol Lett. 2016 Oct;12(4):2782-2788. doi: 10.3892/ol.2016.5012. Epub 2016 Aug 16. Oncol Lett. 2016. PMID: 27698858 Free PMC article.
-
Archetype and Rearranged Non-coding Control Regions in Urothelial Bladder Carcinoma of Immunocompetent Individuals.Cancer Genomics Proteomics. 2016 11-12;13(6):499-509. doi: 10.21873/cgp.20013. Cancer Genomics Proteomics. 2016. PMID: 27807073 Free PMC article.
-
Association between hMLH1 hypermethylation and JC virus (JCV) infection in human colorectal cancer (CRC).Clin Epigenetics. 2011 Apr;2(1):1-5. doi: 10.1007/s13148-010-0013-3. Epub 2010 Nov 24. Clin Epigenetics. 2011. PMID: 22704265 Free PMC article.
-
RNASEL -1385G/A polymorphism and cancer risk: a meta-analysis based on 21 case-control studies.Mol Biol Rep. 2011 Nov;38(8):5099-105. doi: 10.1007/s11033-010-0657-2. Epub 2011 Jan 9. Mol Biol Rep. 2011. PMID: 21221811
-
Presence of JC virus DNA in the tumor tissue and normal mucosa of patients with sporadic colorectal cancer (CRC) or with positive family history and Bethesda criteria.Dig Dis Sci. 2012 Jan;57(1):79-84. doi: 10.1007/s10620-011-1855-z. Epub 2011 Aug 10. Dig Dis Sci. 2012. PMID: 21830098
References
-
- Lengauer C, Kinzler K W, Vogelstein B. Nature (London) 1997;386:623–627. - PubMed
-
- Boland C R, Sato J, Appelman H D, Bresalier R S, Feinberg A P. Nat Med. 1995;1:902–909. - PubMed
-
- Lazutka J R, Neel J V, Major E O, Dedonyte V, Mierauskine J, Slapsyte G, Kesminiene A. Cancer Lett. 1996;109:177–183. - PubMed
-
- Fanning E, Knippers R E. Annu Rev Biochem. 1992;61:55–85. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

