sigmaK can negatively regulate sigE expression by two different mechanisms during sporulation of Bacillus subtilis
- PMID: 10383978
- PMCID: PMC93900
- DOI: 10.1128/JB.181.13.4081-4088.1999
sigmaK can negatively regulate sigE expression by two different mechanisms during sporulation of Bacillus subtilis
Abstract
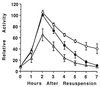
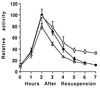
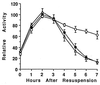
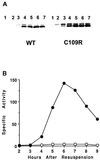
Temporal and spatial gene regulation during Bacillus subtilis sporulation involves the activation and inactivation of multiple sigma subunits of RNA polymerase in a cascade. In the mother cell compartment of sporulating cells, expression of the sigE gene, encoding the earlier-acting sigma factor, sigmaE, is negatively regulated by the later-acting sigma factor, sigmaK. Here, it is shown that the negative feedback loop does not require SinR, an inhibitor of sigE transcription. Production of sigmaK about 1 h earlier than normal does affect Spo0A, which when phosphorylated is an activator of sigE transcription. A mutation in the spo0A gene, which bypasses the phosphorelay leading to the phosphorylation of Spo0A, diminished the negative effect of early sigmaK production on sigE expression early in sporulation. Also, early production of sigmaK reduced expression of other Spo0A-dependent genes but not expression of the Spo0A-independent ald gene. In contrast, both sigE and ald were overexpressed late in development of cells that fail to make sigmaK. The ald promoter, like the sigE promoter, is believed to be recognized by sigmaA RNA polymerase, suggesting that sigmaK may inhibit sigmaA activity late in sporulation. To exert this negative effect, sigmaK must be transcriptionally active. A mutant form of sigmaK that associates with core RNA polymerase, but does not direct transcription of a sigmaK-dependent gene, failed to negatively regulate expression of sigE or ald late in development. On the other hand, the negative effect of early sigmaK production on sigE expression early in sporulation did not require transcriptional activity of sigmaK RNA polymerase. These results demonstrate that sigmaK can negatively regulate sigE expression by two different mechanisms, one observed when sigmaK is produced earlier than normal, which does not require sigmaK to be transcriptionally active and affects Spo0A, and the other observed when sigmaK is produced at the normal time, which requires sigmaK RNA polymerase transcriptional activity. The latter mechanism facilitates the switch from sigmaE to sigmaK in the cascade controlling mother cell gene expression.
Figures



References
-
- Baldus J M, Buckner C M, Moran C P. Evidence that the transcriptional activator Spo0A interacts with two sigma factors in Bacillus subtilis. Mol Microbiol. 1995;17:281–290. - PubMed
-
- Bird T H, Grimsley J K, Hoch J A, Spiegelman G B. The Bacillus subtilis response regulator Spo0A stimulates transcription of the spoIIG operon through modification of RNA polymerase promoter complex. J Mol Biol. 1996;256:436–448. - PubMed
-
- Burbulys D, Trach K A, Hoch J A. Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. Cell. 1991;64:545–552. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

