Assembly of 5S ribosomal RNA is required at a specific step of the pre-rRNA processing pathway
- PMID: 10385518
- PMCID: PMC2133170
- DOI: 10.1083/jcb.145.7.1369
Assembly of 5S ribosomal RNA is required at a specific step of the pre-rRNA processing pathway
Abstract
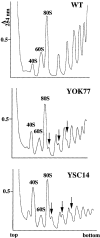
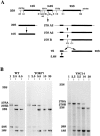
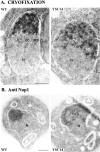
A collection of yeast strains surviving with mutant 5S RNA has been constructed. The mutant strains presented alterations of the nucleolar structure, with less granular component, and a delocalization of the 25S rRNA throughout the nucleoplasm. The 5S RNA mutations affected helix I and resulted in decreased amounts of stable 5S RNA and of the ribosomal 60S subunits. The shortage of 60S subunits was due to a specific defect in the processing of the 27SB precursor RNA that gives rise to the mature 25S and 5.8S rRNA. The processing rate of the 27SB pre-rRNA was specifically delayed, whereas the 27SA and 20S pre-rRNA were processed at a normal rate. The defect was partially corrected by increasing the amount of mutant 5S RNA. We propose that the 5S RNA is recruited by the pre-60S particle and that its recruitment is necessary for the efficient processing of the 27SB RNA precursor. Such a mechanism could ensure that all newly formed mature 60S subunits contain stoichiometric amounts of the three rRNA components.
Figures



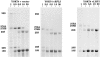
Similar articles
-
Assembly factors Rpf2 and Rrs1 recruit 5S rRNA and ribosomal proteins rpL5 and rpL11 into nascent ribosomes.Genes Dev. 2007 Oct 15;21(20):2580-92. doi: 10.1101/gad.1569307. Genes Dev. 2007. PMID: 17938242 Free PMC article.
-
Rlp7p is associated with 60S preribosomes, restricted to the granular component of the nucleolus, and required for pre-rRNA processing.J Cell Biol. 2002 Jun 10;157(6):941-51. doi: 10.1083/jcb.200111039. Epub 2002 Jun 10. J Cell Biol. 2002. PMID: 12058014 Free PMC article.
-
Ribosomal proteins L7 and L8 function in concert with six A₃ assembly factors to propagate assembly of domains I and II of 25S rRNA in yeast 60S ribosomal subunits.RNA. 2012 Oct;18(10):1805-22. doi: 10.1261/rna.032540.112. Epub 2012 Aug 14. RNA. 2012. PMID: 22893726 Free PMC article.
-
Nuclear export and cytoplasmic maturation of ribosomal subunits.FEBS Lett. 2007 Jun 19;581(15):2783-93. doi: 10.1016/j.febslet.2007.05.013. Epub 2007 May 11. FEBS Lett. 2007. PMID: 17509569 Review.
-
Principles of 60S ribosomal subunit assembly emerging from recent studies in yeast.Biochem J. 2017 Jan 15;474(2):195-214. doi: 10.1042/BCJ20160516. Biochem J. 2017. PMID: 28062837 Free PMC article. Review.
Cited by
-
Association of yeast RNA polymerase I with a nucleolar substructure active in rRNA synthesis and processing.J Cell Biol. 2000 May 1;149(3):575-90. doi: 10.1083/jcb.149.3.575. J Cell Biol. 2000. PMID: 10791972 Free PMC article.
-
Placeholder factors in ribosome biogenesis: please, pave my way.Microb Cell. 2017 Apr 27;4(5):144-168. doi: 10.15698/mic2017.05.572. Microb Cell. 2017. PMID: 28685141 Free PMC article. Review.
-
A novel subunit of yeast RNA polymerase III interacts with the TFIIB-related domain of TFIIIB70.Mol Cell Biol. 2000 Jan;20(2):488-95. doi: 10.1128/MCB.20.2.488-495.2000. Mol Cell Biol. 2000. PMID: 10611227 Free PMC article.
-
Two trypanosome-specific proteins are essential factors for 5S rRNA abundance and ribosomal assembly in Trypanosoma brucei.Eukaryot Cell. 2007 Oct;6(10):1766-72. doi: 10.1128/EC.00119-07. Epub 2007 Aug 22. Eukaryot Cell. 2007. PMID: 17715362 Free PMC article.
-
The essential nucleolar yeast protein Nop8p controls the exosome function during 60S ribosomal subunit maturation.PLoS One. 2011;6(6):e21686. doi: 10.1371/journal.pone.0021686. Epub 2011 Jun 29. PLoS One. 2011. PMID: 21747919 Free PMC article.
References
-
- Allison LA, North MT, Neville LA. Differential binding of oocyte-type and somatic-type 5S rRNA to TFIIIA and ribosomal protein L5 in Xenopus oocytes: specialization for storage versus mobilization. Dev Biol. 1995;168:284–295. - PubMed
-
- Archambault J, Milne CA, Schappert KT, Baum B, Friesen JD, Segall J. The deduced sequence of the transcription factor TFIIIA from Saccharomyces cerevisiae reveals extensive divergence from XenopusTFIIIA. J Biol Chem. 1992;267:3282–3288. - PubMed
-
- Ausubel, F.M., R. Brent, R.E. Kingston, D.D. Moore, J.G. Seidman, J.A. Smith, and K. Struhl. 1987. Current Protocols in Molecular Biology. Vols. 1 and 2. G.P.A. Wiley-Interscience, New York. 1.0.1–16.21.9.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

