ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly
- PMID: 10385622
- PMCID: PMC316812
- DOI: 10.1101/gad.13.12.1529
ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly
Abstract
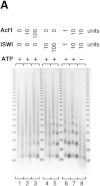
The assembly of core histones and DNA into periodic nucleosome arrays is mediated by ACF, an ISWI-containing factor, and NAP-1, a core histone chaperone, in an ATP-dependent process. We describe the isolation of Drosophila acf1 cDNA, which encodes the p170 and p185 forms of the Acf1 protein in ACF. Acf1 is a novel protein that contains two PHD fingers, one bromodomain, and two new conserved regions. Human WSTF, which is encoded by one of multiple genes that is deleted in Williams syndrome individuals, is the only currently known mammalian protein with each of the conserved motifs in Acf1. Purification of the native form of Acf1 led to the isolation of ACF comprising Acf1 (both p170 and p185 forms) and ISWI. Native Acf1 did not copurify with components of NURF or CHRAC, which are other ISWI-containing complexes in Drosophila. Purified recombinant ACF, consisting of Acf1 (either p185 alone or both p170 and p185) and ISWI, catalyzes the deposition of histones into extended periodic nucleosome arrays. Notably, the Acf1 and ISWI subunits function synergistically in the assembly of chromatin. ISWI alone exhibits a weak activity that is approximately 3% that of ACF. These results indicate that both Acf1 and ISWI participate in the chromatin assembly process and suggest further that the Acf1 subunit confers additional functionality to the general 'motor' activity of ISWI.
Figures










References
-
- Aasland R, Gibson TJ, Stewart AF. The PHD finger: Implications for chromatin-mediated transcriptional regulation. Trends Biochem Sci. 1995;20:56–59. - PubMed
-
- Adams CR, Kamakaka RT. Chromatin assembly: Biochemical identities and genetic redundancy. Curr Opin Genet Dev. 1999;9:185–190. - PubMed
-
- Annunziato AT. Histone acetylation during chromatin replication and nucleosome assembly. In: Wolffe AP, editor. The nucleus. Vol. 1. Greenwich, CT: JAI Press; 1995. pp. 31–56.
-
- Bellugi U, Bihrle A, Jernigan T, Trauner D, Doherty S. Neuropsychological, neurological, and neuroanatomical profile of Williams syndrome. Am J Hum Genet (Suppl.) 1990;6:115–125. - PubMed
-
- Bulger M, Kadonaga JT. Biochemical reconstitution of chromatin with physiological nucleosome spacing. Methods Mol Genet. 1994;5:241–262.
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
