An outbreak of nonflocculating catabolic populations caused the breakdown of a phenol-digesting activated-sludge process
- PMID: 10388669
- PMCID: PMC91422
- DOI: 10.1128/AEM.65.7.2813-2819.1999
An outbreak of nonflocculating catabolic populations caused the breakdown of a phenol-digesting activated-sludge process
Abstract
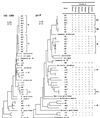
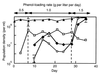
Activated sludge was fed phenol as the sole carbon source, and the phenol-loading rate was increased stepwise from 0.5 to 1.0 g liter-1 day-1 and then to 1.5 g liter-1 day-1. After the loading rate was increased to 1.5 g liter-1 day-1, nonflocculating bacteria outgrew the sludge, and the activated-sludge process broke down within 1 week. The bacterial population structure of the activated sludge was analyzed by temperature gradient gel electrophoresis (TGGE) of PCR-amplified 16S ribosomal DNA (rDNA) fragments. We found that the population diversity decreased as the phenol-loading rate increased and that two populations (designated populations R6 and R10) predominated in the sludge during the last several days before breakdown. The R6 population was present under the low-phenol-loading-rate conditions, while the R10 population was present only after the loading rate was increased to 1.5 g liter-1 day-1. A total of 41 bacterial strains with different repetitive extragenic palindromic sequence PCR patterns were isolated from the activated sludge under different phenol-loading conditions, and the 16S rDNA and gyrB fragments of these strains were PCR amplified and sequenced. Some bacterial isolates could be associated with major TGGE bands by comparing the 16S rDNA sequences. All of the bacterial strains affiliated with the R6 population had almost identical 16S rDNA sequences, while the gyrB phylogenetic analysis divided these strains into two physiologically divergent groups; both of these groups of strains could grow on phenol, while one group (designated the R6F group) flocculated in laboratory media and the other group (the R6T group) did not. A competitive PCR analysis in which specific gyrB sequences were used as the primers showed that a population shift from R6F to R6T occurred following the increase in the phenol-loading rate to 1.5 g liter-1 day-1. The R10 population corresponded to nonflocculating phenol-degrading bacteria. Our results suggest that an outbreak of nonflocculating catabolic populations caused the breakdown of the activated-sludge process. This study also demonstrated the usefulness of gyrB-targeted fine population analyses in microbial ecology.
Figures


References
-
- Amann R, Lemmer H, Wagner M. Monitoring the community structure of wastewater treatment plants: a comparison of old and new techniques. FEMS Microbiol Ecol. 1998;25:205–216.
-
- American Petroleum Institute. Manual on the disposal of refinery wastes. Volume on liquid waste. Washington, D.C: American Petroleum Institute; 1969.
-
- Beltrame P, Beltrame P L, Carniti P, Pitea D. Kinetics of phenol degradation by activated sludge: value of measurements in a batch reactor. Water Res. 1979;13:1305–1309.
-
- Blattner F R, Plunkett G, 3rd, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

