Identification of a new gene family expressed during the onset of sexual reproduction in the centric diatom Thalassiosira weissflogii
- PMID: 10388712
- PMCID: PMC91465
- DOI: 10.1128/AEM.65.7.3121-3128.1999
Identification of a new gene family expressed during the onset of sexual reproduction in the centric diatom Thalassiosira weissflogii
Abstract
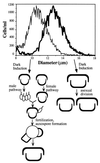
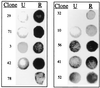
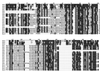
An intriguing feature of the diatom life cycle is that sexual reproduction and the generation of genetic diversity are coupled to the control of cell size. A PCR-based cDNA subtraction technique was used to identify genes that are expressed as small cells of the centric diatom Thalassiosira weissflogii initiate gametogenesis. Ten genes that are up-regulated during the early stages of sexual reproduction have been identified thus far. Three of the sexually induced genes, Sig1, Sig2, and Sig3, were sequenced to completion and are members of a novel gene family. The three polypeptides encoded by these genes possess different molecular masses and charges but display many features in common: they share five highly conserved domains; they each contain three or more cysteine-rich epithelial growth factor (EGF)-like repeats; and they each display homology to the EGF-like region of the vertebrate extracellular matrix glycoprotein tenascin X. Interestingly, the five conserved domains appear in the same order in each polypeptide but are separated by variable numbers of nonconserved amino acids. SIG1 and SIG2 display putative regulatory domains within the nonconserved regions. A calcium-binding, EF-hand motif is found in SIG1, and an ATP/GTP binding motif is present in SIG2. The striking similarity between the SIG polypeptides and extracellular matrix components commonly involved in cell-cell interactions suggests that the SIG polypeptides may play a role in sperm-egg recognition. The SIG polypeptides are thus important molecular targets for determining when and where sexual reproduction occurs in the field.
Figures


References
-
- Armbrust E V, Chisholm S W. Patterns of cell size change in a marine diatom: variability evolving from clonal isolates. J Phycol. 1992;28:146–156.
-
- Armbrust E V, Olson R J, Chisholm S W. Role of light and the cell cycle on the induction of spermatogenesis in a centric diatom. J Phycol. 1990;26:470–478.
-
- Baylor College of Medicine. 3 May 1999, revision date. [Online.] Clustal W. Human Genome Sequencing Center, Baylor College of Medicine, Houston, Tex. http://dot.imgen.bcm.tmc.edu:9331/multi-align/multi-align.html. [20 February 1999, last date accessed.]
-
- Braarud T. Microspores in diatoms. Nature. 1939;143:899.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

