Seasonal dynamics of bacterioplankton community structure in a eutrophic lake as determined by 5S rRNA analysis
- PMID: 10388718
- PMCID: PMC91471
- DOI: 10.1128/AEM.65.7.3164-3174.1999
Seasonal dynamics of bacterioplankton community structure in a eutrophic lake as determined by 5S rRNA analysis
Abstract
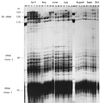
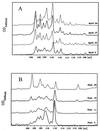
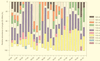
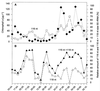
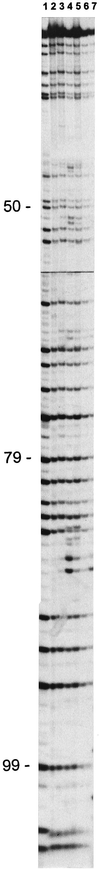
Community structure of bacterioplankton was studied during the major growth season for phytoplankton (April to October) in the epilimnion of a temperate eutrophic lake (Lake Plusssee, northern Germany) by using comparative 5S rRNA analysis. Estimates of the relative abundances of single taxonomic groups were made on the basis of the amounts of single 5S rRNA bands obtained after high-resolution electrophoresis of RNA directly from the bacterioplankton. Full-sequence analysis of single environmental 5S rRNAs enabled the identification of single taxonomic groups of bacteria. Comparison of partial 5S rRNA sequences allowed the detection of changes of single taxa over time. Overall, the whole bacterioplankton community showed two to eight abundant (>4% of the total 5S rRNA) taxa. A distinctive seasonal succession was observed in the taxonomic structure of this pelagic community. A rather-stable community structure, with seven to eight different taxonomic units, was observed beginning in April during the spring phytoplankton bloom. A strong reduction in this diversity occurred at the beginning of the clear-water phase (early May), when only two to four abundant taxa were observed, with one taxon dominating (up to 72% of the total 5S rRNA). The community structure during summer stagnation (June and July) was characterized by frequent changes of different dominating taxa. During late summer, a dinoflagellate bloom (Ceratium hirudinella) occurred, with Comamonas acidovorans (beta-subclass of the class Proteobacteria) becoming the dominant bacterial species (average abundance of 43% of the total 5S rRNA). Finally, the seasonal dynamics of the community structure of bacterioplankton were compared with the abundances of other major groups of the aquatic food web, such as phyto- and zooplankton, revealing that strong grazing pressure by zooplankton can reduce microbial diversity substantially in pelagic environments.
Figures



Similar articles
-
Bacterioplankton community shifts in an arctic lake correlate with seasonal changes in organic matter source.Appl Environ Microbiol. 2003 Apr;69(4):2253-68. doi: 10.1128/AEM.69.4.2253-2268.2003. Appl Environ Microbiol. 2003. PMID: 12676708 Free PMC article.
-
Changes in bacterioplankton community structure and activity with depth in a eutrophic lake as revealed by 5S rRNA analysis.Appl Environ Microbiol. 2002 Jul;68(7):3606-13. doi: 10.1128/AEM.68.7.3606-3613.2002. Appl Environ Microbiol. 2002. PMID: 12089049 Free PMC article.
-
Bacterioplankton community structure and dynamics after large-scale release of nonindigenous bacteria as revealed by low-molecular-weight-RNA analysis.Appl Environ Microbiol. 1992 Oct;58(10):3387-94. doi: 10.1128/aem.58.10.3387-3394.1992. Appl Environ Microbiol. 1992. PMID: 1280060 Free PMC article.
-
Phytoplankton community succession shaping bacterioplankton community composition in Lake Taihu, China.Water Res. 2011 Aug;45(14):4169-82. doi: 10.1016/j.watres.2011.05.022. Epub 2011 May 31. Water Res. 2011. PMID: 21684570
-
Limitations of sequence dissimilarity as a predictor of prokaryotic lineage.Open Biol. 2025 Mar;15(3):240302. doi: 10.1098/rsob.240302. Epub 2025 Mar 19. Open Biol. 2025. PMID: 40101780 Free PMC article. Review.
Cited by
-
A meta-analysis of changes in bacterial and archaeal communities with time.ISME J. 2013 Aug;7(8):1493-506. doi: 10.1038/ismej.2013.54. Epub 2013 Apr 11. ISME J. 2013. PMID: 23575374 Free PMC article.
-
Population dynamics and diversity of viruses, bacteria and phytoplankton in a shallow eutrophic lake.Microb Ecol. 2008 Jul;56(1):29-42. doi: 10.1007/s00248-007-9321-3. Epub 2007 Oct 10. Microb Ecol. 2008. PMID: 17924158 Free PMC article.
-
Bacterioplankton community shifts in an arctic lake correlate with seasonal changes in organic matter source.Appl Environ Microbiol. 2003 Apr;69(4):2253-68. doi: 10.1128/AEM.69.4.2253-2268.2003. Appl Environ Microbiol. 2003. PMID: 12676708 Free PMC article.
-
Changes in bacterioplankton community structure and activity with depth in a eutrophic lake as revealed by 5S rRNA analysis.Appl Environ Microbiol. 2002 Jul;68(7):3606-13. doi: 10.1128/AEM.68.7.3606-3613.2002. Appl Environ Microbiol. 2002. PMID: 12089049 Free PMC article.
-
Geographic and environmental sources of variation in lake bacterial community composition.Appl Environ Microbiol. 2005 Jan;71(1):227-39. doi: 10.1128/AEM.71.1.227-239.2005. Appl Environ Microbiol. 2005. PMID: 15640192 Free PMC article.
References
-
- Azam F, Fenchel T, Field J G, Gray J S, Meyer-Reil L A, Thingstad F. The ecological role of water-column microbes in the sea. Mar Ecol Prog Ser. 1983;10:257–263.
-
- Bemmer A, Overbeck J. Zooplankton grazing on bacteria. In: Overbeck J, Chróst R J, editors. Microbial ecology of Lake Plußsee. New York, N.Y: Springer; 1994. pp. 229–250.
-
- Berninger U-G, Finlay B J, Kuuppo-Leinikki P. Protozoan control of bacterial abundances in freshwater. Limol Oceanogr. 1991;36:139–147.
-
- Boschker H T S, Nold S C, Wellsbury P, Bos D, de Graaf W, Pel R, Parkes R J, Cappenberg T E. Direct linking of microbial populations to specific biogeochemical processes by 13C-labelling of biomarkers. Nature (London) 1998;392:801–804.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

