Identification and analysis of the balhimycin biosynthetic gene cluster and its use for manipulating glycopeptide biosynthesis in Amycolatopsis mediterranei DSM5908
- PMID: 10390204
- PMCID: PMC89325
- DOI: 10.1128/AAC.43.7.1565
Identification and analysis of the balhimycin biosynthetic gene cluster and its use for manipulating glycopeptide biosynthesis in Amycolatopsis mediterranei DSM5908
Abstract
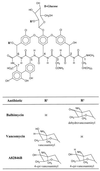
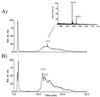
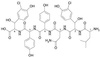
Seven complete genes and one incomplete gene for the biosynthesis of the glycopeptide antibiotic balhimycin were isolated from the producer, Amycolatopsis mediterranei DSM5908, by a reverse-cloning approach and characterized. Using oligonucleotides derived from glycosyltransferase sequences, a 900-bp glycosyltransferase gene fragment was amplified and used to identify a DNA fragment of 9,882 bp. Of the identified open reading frames, three (oxyA to -C) showed significant sequence similarities to cytochrome P450 monooxygenases and one (bhaA) showed similarities to halogenase, and the genes bgtfA to -C showed similarities to glycosyltransferases. Glycopeptide biosynthetic mutants were created by gene inactivation experiments eliminating oxygenase and glycosyltransferase functions. Inactivation of the oxygenase gene(s) resulted in a balhimycin mutant (SP1-1) which was not able to synthesize an antibiotically active compound. Structural analysis by high-performance liquid chromatography-mass spectrometry, fragmentation studies, and amino acid analysis demonstrated that these oxygenases are involved in the coupling of the aromatic side chains of the unusual heptapeptide. Mutant strain HD1, created by inactivation of the glycosyltransferase gene bgtfB, produced at least four different compounds which were not glycosylated but still antibiotically active.
Figures



Similar articles
-
Glycopeptide biosynthesis in Amycolatopsis mediterranei DSM5908: function of a halogenase and a haloperoxidase/perhydrolase.Chem Biol. 2002 Feb;9(2):225-35. doi: 10.1016/s1074-5521(02)00101-1. Chem Biol. 2002. PMID: 11880037
-
Genetic analysis of the balhimycin (vancomycin-type) oxygenase genes.J Biotechnol. 2006 Aug 5;124(4):640-53. doi: 10.1016/j.jbiotec.2006.04.009. Epub 2006 May 30. J Biotechnol. 2006. PMID: 16730832
-
Cloning and analysis of a peptide synthetase gene of the balhimycin producer Amycolatopsis mediterranei DSM5908 and development of a gene disruption/replacement system.J Biotechnol. 1997 Aug 11;56(2):115-28. doi: 10.1016/s0168-1656(97)00082-5. J Biotechnol. 1997. PMID: 9304873
-
The biosynthesis of glycopeptide antibiotics--a model for complex, non-ribosomally synthesized, peptidic secondary metabolites.Appl Microbiol Biotechnol. 2004 Jan;63(4):344-50. doi: 10.1007/s00253-003-1443-z. Epub 2003 Oct 15. Appl Microbiol Biotechnol. 2004. PMID: 14564489 Review.
-
Combinatorial glycosylation of glycopeptide antibiotics.Chem Biol. 2002 Dec;9(12):1268-70. doi: 10.1016/s1074-5521(02)00289-2. Chem Biol. 2002. PMID: 12498878 Review.
Cited by
-
Comparison of Antibiotic Resistance Mechanisms in Antibiotic-Producing and Pathogenic Bacteria.Molecules. 2019 Sep 21;24(19):3430. doi: 10.3390/molecules24193430. Molecules. 2019. PMID: 31546630 Free PMC article. Review.
-
Differential proteomic analysis highlights metabolic strategies associated with balhimycin production in Amycolatopsis balhimycina chemostat cultivations.Microb Cell Fact. 2010 Nov 26;9:95. doi: 10.1186/1475-2859-9-95. Microb Cell Fact. 2010. PMID: 21110849 Free PMC article.
-
Detection, distribution, and organohalogen compound discovery implications of the reduced flavin adenine dinucleotide-dependent halogenase gene in major filamentous actinomycete taxonomic groups.Appl Environ Microbiol. 2009 Jul;75(14):4813-20. doi: 10.1128/AEM.02958-08. Epub 2009 May 15. Appl Environ Microbiol. 2009. PMID: 19447951 Free PMC article.
-
Chemoenzymatic and template-directed synthesis of bioactive macrocyclic peptides.Microbiol Mol Biol Rev. 2006 Mar;70(1):121-46. doi: 10.1128/MMBR.70.1.121-146.2006. Microbiol Mol Biol Rev. 2006. PMID: 16524919 Free PMC article. Review.
-
Genome mining reveals the genus Xanthomonas to be a promising reservoir for new bioactive non-ribosomally synthesized peptides.BMC Genomics. 2013 Sep 27;14:658. doi: 10.1186/1471-2164-14-658. BMC Genomics. 2013. PMID: 24069909 Free PMC article.
References
-
- Arthur M, Depardieu F, Reynolds P, Courvalin P. Quantitative analysis of the metabolism of soluble cytoplasmic peptidoglycan precursors of glycopeptide-resistant enterococci. Mol Microbiol. 1996;21:33–44. - PubMed
-
- Bierman M, Logan R, O’Brien K, Seno E T, Rao R N, Schoner B E. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene. 1992;116:43–49. - PubMed
-
- Bullock W O, Fernandez J M, Short J M. XL1-Blue, a high efficiency plasmid transforming recA Escherichia coli strain with beta galactosidase selection. Focus. 1987;5:376–378.
-
- Chatterjee S, Vijayakumar E K S, Nadkarni S R, Patel M V, Blumbach J, Ganguli B N. Balhimycin, a new glycopeptide antibiotic with an unusual hydrated 3-amino-4-oxoaldopyranose sugar moiety. J Org Chem. 1994;59:3480–3484.
-
- Eggink G, Engel H, Vriend G, Terpstra P, Witholt B. Rubredoxin reductase of Pseudomonas oleovorans: structural relationship to other flavoprotein oxidoreductases based on one NAD and two FAD fingerprints. J Mol Biol. 1990;212:135–142. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

