The myeloperoxidase system of human phagocytes generates Nepsilon-(carboxymethyl)lysine on proteins: a mechanism for producing advanced glycation end products at sites of inflammation
- PMID: 10393704
- PMCID: PMC408396
- DOI: 10.1172/JCI3042
The myeloperoxidase system of human phagocytes generates Nepsilon-(carboxymethyl)lysine on proteins: a mechanism for producing advanced glycation end products at sites of inflammation
Abstract
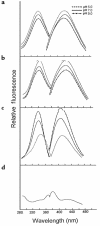
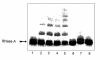
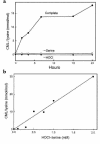
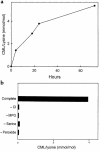
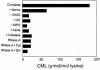
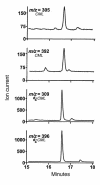
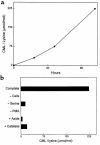
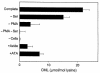
Reactive aldehydes derived from reducing sugars and peroxidation of lipids covalently modify proteins and may contribute to oxidative tissue damage. We recently described another mechanism for generating reactive aldehydes from free alpha-amino acids. The pathway begins with myeloperoxidase, a heme enzyme secreted by activated neutrophils. Conversion of alpha-amino acids to aldehydes requires hypochlorous acid (HOCl), formed from H2O2 and chloride by myeloperoxidase. When L-serine is the substrate, HOCl generates high yields of glycolaldehyde. We now demonstrate that a model protein, ribonuclease A (RNase A), exposed to free L-serine and HOCl exhibits the biochemical hallmarks of advanced glycation end (AGE) products -- browning, increased fluorescence, and cross-linking. Furthermore, Nepsilon-(carboxymethyl)lysine (CML), a chemically well-characterized AGE product, was generated on RNase A when it was exposed to reagent HOCl-serine, the myeloperoxidase-H2O2-chloride system plus L-serine, or activated human neutrophils plus L-serine. CML production by neutrophils was inhibited by the H2O2 scavenger catalase and the heme poison azide, implicating myeloperoxidase in the cell-mediated reaction. CML was also generated on RNase A by a myeloperoxidase-dependent pathway when neutrophils were activated in a mixture of amino acids. Under these conditions, we observed both L-serine-dependent and L-serine-independent pathways of CML formation. The in vivo production of glycolaldehyde and other reactive aldehydes by myeloperoxidase may thus play an important pathogenic role by generating AGE products and damaging tissues at sites of inflammation.
Figures

References
-
- Esterbauer HR, Schaur J, Zollner H. Chemistry and biochemistry of 4-hydroxynonenal, malondialdehyde and related aldehydes. Free Radic Biol Med. 1991;11:81–128. - PubMed
-
- Baynes JW. Role of oxidative stress in development of complications in diabetes. Diabetes. 1991;40:405–412. - PubMed
-
- Monnier VM, Sell DR, Nagaraj RH, Miyata S. Mechanisms of protection against damage mediated by the Maillard reaction in aging. Gerontology. 1991;37:152–165. - PubMed
-
- Berliner JA, Heinecke JW. The role of oxidized lipoproteins in atherogenesis. Free Radic Biol Med. 1996;20:707–727. - PubMed
-
- Witz G. Biological interactions of alpha, beta-unsaturated aldehydes. Free Radic Biol Med. 1989;7:333–349. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

