RecA polymerization on double-stranded DNA by using single-molecule manipulation: the role of ATP hydrolysis
- PMID: 10393922
- PMCID: PMC22162
- DOI: 10.1073/pnas.96.14.7916
RecA polymerization on double-stranded DNA by using single-molecule manipulation: the role of ATP hydrolysis
Abstract
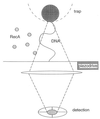
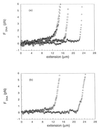
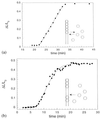
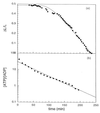
The polymerization of RecA on individual double-stranded DNA molecules is studied. A linear DNA (lambda DNA, 48.5 Kb), anchored at one end to a cover glass and at the other end to an optically trapped 3-micrometers diameter polystyrene bead, serves as a template. The elongation caused by RecA assembly is measured in the presence of ATP and ATP[gammaS]. By using force extension and hydrodynamic recoil, a value of the persistence length of the RecA-DNA complex is obtained. In the presence of ATP, the polymer length is unstable, first growing to saturation and then decreasing. This suggests a transient dynamics of association and dissociation for RecA on a double-stranded DNA, the process being controlled by ATP hydrolysis. Part of this dynamics is suppressed in the presence of ATP[gammaS], leading to a stabilized RecA-DNA complex. A one-dimensional nucleation and growth model is presented that may account for the protein assembly.
Figures


References
-
- Kowalczykowski S C, Eggleston A K. Annu Rev Biochem. 1994;63:991–1043. - PubMed
-
- Roca A I, Cox M M. Crit Rev Biochem Mol Biol. 1990;25:415–456. - PubMed
-
- Cox M M. Trends Biochem Sci. 1994;19:217–222. - PubMed
-
- Stasiak A, Di Capua E, Koller T. J Mol Biol. 1981;151:557–564. - PubMed
-
- Stasiak A, Di Capua E. Nature (London) 1982;299:185–186. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

