The nop-1 gene of Neurospora crassa encodes a seven transmembrane helix retinal-binding protein homologous to archaeal rhodopsins
- PMID: 10393943
- PMCID: PMC22183
- DOI: 10.1073/pnas.96.14.8034
The nop-1 gene of Neurospora crassa encodes a seven transmembrane helix retinal-binding protein homologous to archaeal rhodopsins
Abstract
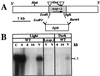
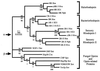
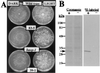
Opsins are a class of retinal-binding, seven transmembrane helix proteins that function as light-responsive ion pumps or sensory receptors. Previously, genes encoding opsins had been identified in animals and the Archaea but not in fungi or other eukaryotic microorganisms. Here, we report the identification and mutational analysis of an opsin gene, nop-1, from the eukaryotic filamentous fungus Neurospora crassa. The nop-1 amino acid sequence predicts a protein that shares up to 81.8% amino acid identity with archaeal opsins in the 22 retinal binding pocket residues, including the conserved lysine residue that forms a Schiff base linkage with retinal. Evolutionary analysis revealed relatedness not only between NOP-1 and archaeal opsins but also between NOP-1 and several fungal opsin-related proteins that lack the Schiff base lysine residue. The results provide evidence for a eukaryotic opsin family homologous to the archaeal opsins, providing a plausible link between archaeal and visual opsins. Extensive analysis of Deltanop-1 strains did not reveal obvious defects in light-regulated processes under normal laboratory conditions. However, results from Northern analysis support light and conidiation-based regulation of nop-1 gene expression, and NOP-1 protein heterologously expressed in Pichia pastoris is labeled by using all-trans [3H]retinal, suggesting that NOP-1 functions as a rhodopsin in N. crassa photobiology.
Figures

References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

