Structural basis of chaperone self-capping in P pilus biogenesis
- PMID: 10393968
- PMCID: PMC22208
- DOI: 10.1073/pnas.96.14.8178
Structural basis of chaperone self-capping in P pilus biogenesis
Abstract
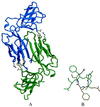
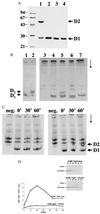
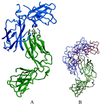
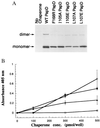
PapD is an immunoglobulin-like chaperone that mediates the assembly of P pili in uropathogenic strains of Escherichia coli. It binds and caps interactive surfaces on pilus subunits to prevent their premature associations in the periplasm. We elucidated the structural basis of a mechanism whereby PapD also interacts with itself, capping its own subunit binding surface. Crystal structures of dimeric forms of PapD revealed that this self-capping mechanism involves a rearrangement and ordering of the C2-D2 and F1-G1 loops upon dimerization which might ensure that a stable dimer is not formed in solution in spite of a relatively large dimer interface. An analysis of site directed mutations revealed that chaperone dimerization requires the same surface that is otherwise used to bind subunits.
Figures
References
-
- Normark S, Baga M, Goransson M, Lindberg F P, Lund B, Norgren M, Uhlin B E. In: Microbial Lectins and Agglutinins: Properties and Biological Activity. Mirelman D, editor. New York: Wiley Interscience; 1986. pp. 113–143.
-
- Hultgren S J, Abraham S N, Caparon M G, Falk P, St. Geme J W, III, Normark S. Cell. 1993;73:887–901. - PubMed
-
- Kuehn M J, Heuser J, Normark S, Hultgren S J. Nature (London) 1992;356:252–255. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

