The plant vesicle-associated SNARE AtVTI1a likely mediates vesicle transport from the trans-Golgi network to the prevacuolar compartment
- PMID: 10397763
- PMCID: PMC25440
- DOI: 10.1091/mbc.10.7.2251
The plant vesicle-associated SNARE AtVTI1a likely mediates vesicle transport from the trans-Golgi network to the prevacuolar compartment
Abstract
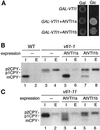
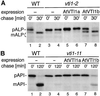
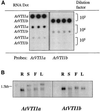
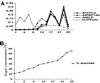
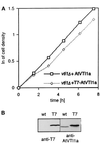
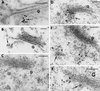
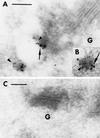
Membrane traffic in eukaryotic cells relies on recognition between v-SNAREs on transport vesicles and t-SNAREs on target membranes. Here we report the identification of AtVTI1a and AtVTI1b, two Arabidopsis homologues of the yeast v-SNARE Vti1p, which is required for multiple transport steps in yeast. AtVTI1a and AtVTI1b share 60% amino acid identity with one another and are 32 and 30% identical to the yeast protein, respectively. By suppressing defects found in specific strains of yeast vti1 temperature-sensitive mutants, we show that AtVTI1a can substitute for Vti1p in Golgi-to-prevacuolar compartment (PVC) transport, whereas AtVTI1b substitutes in two alternative pathways: the vacuolar import of alkaline phosphatase and the so-called cytosol-to-vacuole pathway used by aminopeptidase I. Both AtVTI1a and AtVTI1b are expressed in all major organs of Arabidopsis. Using subcellular fractionation and immunoelectron microscopy, we show that AtVTI1a colocalizes with the putative vacuolar cargo receptor AtELP on the trans-Golgi network and the PVC. AtVTI1a also colocalizes with the t-SNARE AtPEP12p to the PVC. In addition, AtVTI1a and AtPEP12p can be coimmunoprecipitated from plant cell extracts. We propose that AtVTI1a functions as a v-SNARE responsible for targeting AtELP-containing vesicles from the trans-Golgi network to the PVC, and that AtVTI1b is involved in a different membrane transport process.
Figures



Similar articles
-
AtVPS45 complex formation at the trans-Golgi network.Mol Biol Cell. 2000 Jul;11(7):2251-65. doi: 10.1091/mbc.11.7.2251. Mol Biol Cell. 2000. PMID: 10888666 Free PMC article.
-
The Saccharomyces cerevisiae v-SNARE Vti1p is required for multiple membrane transport pathways to the vacuole.Mol Biol Cell. 1999 Jun;10(6):1719-32. doi: 10.1091/mbc.10.6.1719. Mol Biol Cell. 1999. PMID: 10359592 Free PMC article.
-
A putative vacuolar cargo receptor partially colocalizes with AtPEP12p on a prevacuolar compartment in Arabidopsis roots.Proc Natl Acad Sci U S A. 1998 Aug 18;95(17):9920-5. doi: 10.1073/pnas.95.17.9920. Proc Natl Acad Sci U S A. 1998. PMID: 9707576 Free PMC article.
-
Multiple sorting pathways between the late Golgi and the vacuole in yeast.Biochim Biophys Acta. 1998 Aug 14;1404(1-2):211-30. doi: 10.1016/s0167-4889(98)00058-5. Biochim Biophys Acta. 1998. PMID: 9714809 Review.
-
Targeting of tonoplast proteins to the vacuole.Plant Sci. 2013 Oct;211:132-6. doi: 10.1016/j.plantsci.2013.07.005. Epub 2013 Jul 18. Plant Sci. 2013. PMID: 23987818 Review.
Cited by
-
CLV3 is localized to the extracellular space, where it activates the Arabidopsis CLAVATA stem cell signaling pathway.Plant Cell. 2002 May;14(5):969-77. doi: 10.1105/tpc.002196. Plant Cell. 2002. PMID: 12034890 Free PMC article.
-
Loss-of-function mutations of retromer large subunit genes suppress the phenotype of an Arabidopsis zig mutant that lacks Qb-SNARE VTI11.Plant Cell. 2010 Jan;22(1):159-72. doi: 10.1105/tpc.109.069294. Epub 2010 Jan 19. Plant Cell. 2010. PMID: 20086190 Free PMC article.
-
The Arabidopsis Rab GTPase RabA4b localizes to the tips of growing root hair cells.Plant Cell. 2004 Jun;16(6):1589-603. doi: 10.1105/tpc.021634. Epub 2004 May 21. Plant Cell. 2004. PMID: 15155878 Free PMC article.
-
Creeping Stem 1 regulates directional auxin transport for lodging resistance in soybean.Plant Biotechnol J. 2025 Feb;23(2):377-394. doi: 10.1111/pbi.14503. Epub 2024 Nov 13. Plant Biotechnol J. 2025. PMID: 39535932 Free PMC article.
-
NPSN11 is a cell plate-associated SNARE protein that interacts with the syntaxin KNOLLE.Plant Physiol. 2002 Jun;129(2):530-9. doi: 10.1104/pp.003970. Plant Physiol. 2002. PMID: 12068098 Free PMC article.
References
-
- Advani RJ, Bae HR, Bock JB, Chao DS, Doung YC, Prekeris R, Yoo JS, Scheller RH. Seven novel mammalian SNARE proteins localize to distinct membrane compartments. J Biol Chem. 1998;273:10317–10324. - PubMed
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

