Muscle LIM proteins are associated with muscle sarcomeres and require dMEF2 for their expression during Drosophila myogenesis
- PMID: 10397768
- PMCID: PMC25449
- DOI: 10.1091/mbc.10.7.2329
Muscle LIM proteins are associated with muscle sarcomeres and require dMEF2 for their expression during Drosophila myogenesis
Abstract
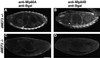
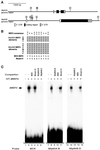
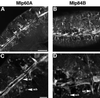
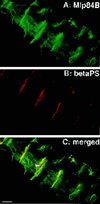
A genetic hierarchy of interactions, involving myogenic regulatory factors of the MyoD and myocyte enhancer-binding 2 (MEF2) families, serves to elaborate and maintain the differentiated muscle phenotype through transcriptional regulation of muscle-specific target genes. Much work suggests that members of the cysteine-rich protein (CRP) family of LIM domain proteins also play a role in muscle differentiation; however, the specific functions of CRPs in this process remain undefined. Previously, we characterized two members of the Drosophila CRP family, the muscle LIM proteins Mlp60A and Mlp84B, which show restricted expression in differentiating muscle lineages. To extend our analysis of Drosophila Mlps, we characterized the expression of Mlps in mutant backgrounds that disrupt specific aspects of muscle development. We show a genetic requirement for the transcription factor dMEF2 in regulating Mlp expression and an ability of dMEF2 to bind, in vitro, to consensus MEF2 sites derived from those present in Mlp genomic sequences. These data suggest that the Mlp genes may be direct targets of dMEF2 within the genetic hierarchy controlling muscle differentiation. Mutations that disrupt myoblast fusion fail to affect Mlp expression. In later stages of myogenic differentiation, which are dedicated primarily to assembly of the contractile apparatus, we analyzed the subcellular distribution of Mlp84B in detail. Immunofluorescent studies revealed the localization of Mlp84B to muscle attachment sites and the periphery of Z-bands of striated muscle. Analysis of mutations that affect expression of integrins and alpha-actinin, key components of these structures, also failed to perturb Mlp84B distribution. In conclusion, we have used molecular epistasis analysis to position Mlp function downstream of events involving mesoderm specification and patterning and concomitant with terminal muscle differentiation. Furthermore, our results are consistent with a structural role for Mlps as components of muscle cytoarchitecture.
Figures





Similar articles
-
Two muscle-specific LIM proteins in Drosophila.J Cell Biol. 1996 Sep;134(5):1179-95. doi: 10.1083/jcb.134.5.1179. J Cell Biol. 1996. PMID: 8794860 Free PMC article.
-
Drosophila melanogaster muscle LIM protein and alpha-actinin function together to stabilize muscle cytoarchitecture: a potential role for Mlp84B in actin-crosslinking.Cytoskeleton (Hoboken). 2013 Jun;70(6):304-16. doi: 10.1002/cm.21106. Epub 2013 Apr 18. Cytoskeleton (Hoboken). 2013. PMID: 23606669 Free PMC article.
-
Different levels, but not different isoforms, of the Drosophila transcription factor DMEF2 affect distinct aspects of muscle differentiation.Dev Biol. 1999 Nov 1;215(1):130-45. doi: 10.1006/dbio.1999.9449. Dev Biol. 1999. PMID: 10525355
-
Synergistic up-regulation of muscle LIM protein expression in C2C12 and NIH3T3 cells by myogenin and MEF2C.Mol Genet Genomics. 2009 Jan;281(1):1-10. doi: 10.1007/s00438-008-0393-7. Epub 2008 Nov 6. Mol Genet Genomics. 2009. PMID: 18987887 Review.
-
Muscle development. Making Drosophila muscle.Curr Biol. 1995 Jul 1;5(7):740-2. doi: 10.1016/s0960-9822(95)00149-7. Curr Biol. 1995. PMID: 7583119 Review.
Cited by
-
A novel muscle LIM-only protein is generated from the paxillin gene locus in Drosophila.EMBO Rep. 2001 Sep;2(9):814-20. doi: 10.1093/embo-reports/kve178. Epub 2001 Aug 23. EMBO Rep. 2001. PMID: 11520860 Free PMC article.
-
The role of Drosophila Lamin C in muscle function and gene expression.Development. 2010 Sep;137(18):3067-77. doi: 10.1242/dev.048231. Epub 2010 Aug 11. Development. 2010. PMID: 20702563 Free PMC article.
-
Many P-element insertions affect wing shape in Drosophila melanogaster.Genetics. 2005 Mar;169(3):1461-75. doi: 10.1534/genetics.104.027748. Epub 2004 Nov 15. Genetics. 2005. PMID: 15545659 Free PMC article.
-
Cardiac gene regulatory networks in Drosophila.Biochim Biophys Acta. 2009 Apr;1789(4):343-53. doi: 10.1016/j.bbagrm.2008.09.002. Epub 2008 Sep 24. Biochim Biophys Acta. 2009. PMID: 18849017 Free PMC article. Review.
-
mef2 activity levels differentially affect gene expression during Drosophila muscle development.Proc Natl Acad Sci U S A. 2008 Jan 22;105(3):918-23. doi: 10.1073/pnas.0711255105. Epub 2008 Jan 15. Proc Natl Acad Sci U S A. 2008. PMID: 18198273 Free PMC article.
References
-
- Arber S, Caroni P. Specificity of single LIM motifs in targeting and LIM/LIM interactions in situ. Genes & Dev. 1996;10:289–300. - PubMed
-
- Arber S, Halder G, Caroni P. Muscle LIM protein, a novel essential regulator of myogenesis, promotes myogenic differentiation. Cell. 1994;79:221–231. - PubMed
-
- Arber S, Hunter JJ, Ross J, Jr, Hongo M, Sansig G, Borg J, Perriad J-C, Chien KR, Caroni P. MLP-deficient mice exhibit a disruption of cardiac cytoarchitectural organization, dilated cardiomyopathy, and heart failure. Cell. 1997;88:393–403. - PubMed
-
- Bate M. The embryonic development of larval muscles in Drosophila. Development. 1990;110:791–804. - PubMed
-
- Bockholt SM, Otey CA, Glenny JRJ, Burridge K. Localization of a 215 kDa tyrosine phosphorylated protein that cross-reacts with tensin antibodies. Exp Cell Res. 1992;203:39–46. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

