Yeast Dam1p is required to maintain spindle integrity during mitosis and interacts with the Mps1p kinase
- PMID: 10397771
- PMCID: PMC25456
- DOI: 10.1091/mbc.10.7.2377
Yeast Dam1p is required to maintain spindle integrity during mitosis and interacts with the Mps1p kinase
Abstract
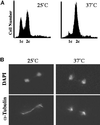
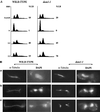
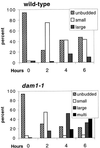
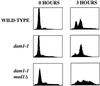
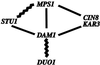
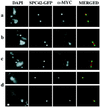
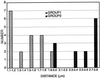
We have identified a mutant allele of the DAM1 gene in a screen for mutations that are lethal in combination with the mps1-1 mutation. MPS1 encodes an essential protein kinase that is required for duplication of the spindle pole body and for the spindle assembly checkpoint. Mutations in six different genes were found to be lethal in combination with mps1-1, of which only DAM1 was novel. The remaining genes encode a checkpoint protein, Bub1p, and four chaperone proteins, Sti1p, Hsc82p, Cdc37p, and Ydj1p. DAM1 is an essential gene that encodes a protein recently described as a member of a microtubule binding complex. We report here that cells harboring the dam1-1 mutation fail to maintain spindle integrity during anaphase at the restrictive temperature. Consistent with this phenotype, DAM1 displays genetic interactions with STU1, CIN8, and KAR3, genes encoding proteins involved in spindle function. We have observed that a Dam1p-Myc fusion protein expressed at endogenous levels and localized by immunofluorescence microscopy, appears to be evenly distributed along short mitotic spindles but is found at the spindle poles at later times in mitosis.
Figures


Similar articles
-
The Saccharomyces cerevisiae spindle pole body duplication gene MPS1 is part of a mitotic checkpoint.J Cell Biol. 1996 Jan;132(1-2):111-23. doi: 10.1083/jcb.132.1.111. J Cell Biol. 1996. PMID: 8567717 Free PMC article.
-
Activation of the budding yeast spindle assembly checkpoint without mitotic spindle disruption.Science. 1996 Aug 16;273(5277):953-6. doi: 10.1126/science.273.5277.953. Science. 1996. PMID: 8688079
-
The yeast protein kinase Mps1p is required for assembly of the integral spindle pole body component Spc42p.J Cell Biol. 2002 Feb 4;156(3):453-65. doi: 10.1083/jcb.200111025. Epub 2002 Feb 4. J Cell Biol. 2002. PMID: 11827982 Free PMC article.
-
Dam1 is the right one: phosphoregulation of kinetochore biorientation.Dev Cell. 2002 Nov;3(5):610-1. doi: 10.1016/s1534-5807(02)00332-5. Dev Cell. 2002. PMID: 12431367 Review.
-
Formin' the connection between microtubules and the cell cortex.J Cell Biol. 1999 Mar 8;144(5):809-11. doi: 10.1083/jcb.144.5.809. J Cell Biol. 1999. PMID: 10085282 Free PMC article. Review. No abstract available.
Cited by
-
The yeast S phase checkpoint enables replicating chromosomes to bi-orient and restrain spindle extension during S phase distress.J Cell Biol. 2005 Mar 28;168(7):999-1012. doi: 10.1083/jcb.200412076. J Cell Biol. 2005. PMID: 15795314 Free PMC article.
-
The highly conserved Ndc80 complex is required for kinetochore assembly, chromosome congression, and spindle checkpoint activity.Genes Dev. 2003 Jan 1;17(1):101-14. doi: 10.1101/gad.1040903. Genes Dev. 2003. PMID: 12514103 Free PMC article.
-
The Hsp70-Hsp90 go-between Hop/Stip1/Sti1 is a proteostatic switch and may be a drug target in cancer and neurodegeneration.Cell Mol Life Sci. 2021 Dec;78(23):7257-7273. doi: 10.1007/s00018-021-03962-z. Epub 2021 Oct 22. Cell Mol Life Sci. 2021. PMID: 34677645 Free PMC article. Review.
-
An alpha-tubulin mutant demonstrates distinguishable functions among the spindle assembly checkpoint genes in Saccharomyces cerevisiae.Genetics. 2002 Jul;161(3):983-94. doi: 10.1093/genetics/161.3.983. Genetics. 2002. PMID: 12136005 Free PMC article.
-
Yeast Dam1p has a role at the kinetochore in assembly of the mitotic spindle.Proc Natl Acad Sci U S A. 2001 Nov 20;98(24):13675-80. doi: 10.1073/pnas.241417098. Epub 2001 Nov 6. Proc Natl Acad Sci U S A. 2001. PMID: 11698664 Free PMC article.
References
-
- Boeke JD, Trueheart J, Natsoulis G, Fink GR. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. - PubMed
-
- Botstein D, Amberg D, Mulholland J, Huffaker T, Adams A, Drubin D, Stearns T. The yeast cytoskeleton. In: Pringle JR, Broach JR, Jones EW, editors. The Molecular and Cellular Biology of the Yeast Saccharomyces: Cell Cycle and Cell Biology. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 1–90.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

