Molecular cloning and tissue-specific expression of an anionic peroxidase in zucchini
- PMID: 10398715
- PMCID: PMC59318
- DOI: 10.1104/pp.120.3.799
Molecular cloning and tissue-specific expression of an anionic peroxidase in zucchini
Abstract
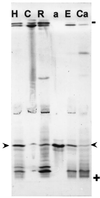
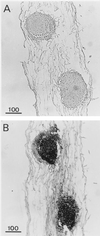
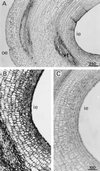
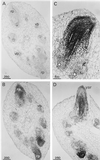
A calcium-pectate-binding anionic isoperoxidase (APRX) from zucchini (Cucurbita pepo) was purified and subjected to N-terminal amino acid microsequencing. The cDNA encoding this enzyme was obtained by reverse transcriptase polymerase chain reaction from a cDNA library. It encoded a mature protein of 309 amino acids exhibiting all of the sequence characteristics of a plant peroxidase. Despite the presence of a C-terminal propeptide, APRX was found in the apoplast. APRX protein and mRNA were found in the root, hypocotyls, and cotyledons. In situ hybridization showed that the APRX-encoding gene was expressed in many different tissues. The strongest expression was observed in root epidermis and in some cells of the stele, in differentiating tracheary elements of hypocotyl, in the lower and upper epidermis, in the palisade parenchyma of cotyledons, and in lateral and adventitious root primordia. In the hypocotyl hook there was an asymmetric expression, with the inner part containing more transcripts than the outer part. Treatment with 2,3,5-triiodobenzoic acid reduced the expression of the APRX-encoding gene in the lower part of the hypocotyl. Our observations suggest that APRX could be involved in lignin formation and that the transcription of its gene was related to auxin level.
Figures







References
-
- Blom TJM, Sierra M, van Vliet TB, Franke-van Dijk MEI, de Koning P, van Iren F, Verpoorte R, Libbenga KR. Uptake and accumulation of ajmalicine into isolated vacuoles of cultured cells of Catharanthus roseus (L.) G. Don. and its conversion into serpentine. Planta. 1991;183:170–177. - PubMed
-
- Catesson A-M, Imberty A, Goldberg R, Czaninski Y (1986) Nature, localization and specificity of peroxidases involved in lignification processes. In H Greppin, C Penel, T Gaspar, eds, Molecular and Physiological Aspects of Plant Peroxidases. University of Geneva, Switzerland, pp 189–198
-
- Davies PJ. Plant Hormones and their Role in Plant Growth and Development. Dordrecht, The Netherlands: Martinus Nijhoff Publishers; 1987.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

