Interaction of Bacillus subtilis Fur (ferric uptake repressor) with the dhb operator in vitro and in vivo
- PMID: 10400588
- PMCID: PMC93932
- DOI: 10.1128/JB.181.14.4299-4307.1999
Interaction of Bacillus subtilis Fur (ferric uptake repressor) with the dhb operator in vitro and in vivo
Abstract
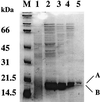
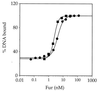
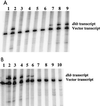
Bacillus subtilis contains three metalloregulatory proteins belonging to the ferric uptake repressor (Fur) family: Fur, Zur, and PerR. We have overproduced and purified Fur protein and analyzed its interaction with the operator region controlling the expression of the dihydroxybenzoate siderophore biosynthesis (dhb) operon. The purified protein binds with high affinity and selectivity to the dhb regulatory region. DNA binding does not require added iron, nor is binding reduced by dialysis of Fur against EDTA or treatment with Chelex. Fur selectively inhibits transcription from the dhb promoter by sigmaA RNA polymerase, even if Fur is added after RNA polymerase holoenzyme. Since neither DNA binding nor inhibition of transcription requires the addition of ferrous ion in vitro, the mechanism by which iron regulates Fur function in vivo is not obvious. Mutagenesis of the fur gene reveals that in vivo repression of the dhb operon by iron requires His97, a residue thought to be involved in iron sensing in other Fur homologs. Moreover, we identify His96 as a second likely iron ligand, since a His96Ala mutant mediates repression at 50 microM but not at 5 microM iron. Our data lead us to suggest that Fur is able to bind DNA independently of bound iron and that the in vivo role of iron is to counteract the effect of an inhibitory factor, perhaps another metal ion, that antagonizes this DNA-binding activity.
Figures

Similar articles
-
Bacillus subtilis contains multiple Fur homologues: identification of the iron uptake (Fur) and peroxide regulon (PerR) repressors.Mol Microbiol. 1998 Jul;29(1):189-98. doi: 10.1046/j.1365-2958.1998.00921.x. Mol Microbiol. 1998. PMID: 9701813
-
Identification of a zinc-specific metalloregulatory protein, Zur, controlling zinc transport operons in Bacillus subtilis.J Bacteriol. 1998 Nov;180(22):5815-21. doi: 10.1128/JB.180.22.5815-5821.1998. J Bacteriol. 1998. PMID: 9811636 Free PMC article.
-
Molecular characterization of the ferric-uptake regulator, fur, from Staphylococcus aureus.Microbiology (Reading). 2000 Mar;146 ( Pt 3):659-668. doi: 10.1099/00221287-146-3-659. Microbiology (Reading). 2000. PMID: 10746769
-
Transcriptional regulation by Ferric Uptake Regulator (Fur) in pathogenic bacteria.Front Cell Infect Microbiol. 2013 Oct 2;3:59. doi: 10.3389/fcimb.2013.00059. eCollection 2013. Front Cell Infect Microbiol. 2013. PMID: 24106689 Free PMC article. Review.
-
Mechanism and regulation of synthesis of aerobactin in Escherichia coli K12 (pColV-K30).Can J Microbiol. 1992 Jul;38(7):728-33. doi: 10.1139/m92-119. Can J Microbiol. 1992. PMID: 1393837 Review.
Cited by
-
Recognition of DNA by three ferric uptake regulator (Fur) homologs in Bacillus subtilis.J Bacteriol. 2003 Nov;185(21):6348-57. doi: 10.1128/JB.185.21.6348-6357.2003. J Bacteriol. 2003. PMID: 14563870 Free PMC article.
-
Bacillus subtilis Fur represses one of two paralogous haem-degrading monooxygenases.Microbiology (Reading). 2011 Nov;157(Pt 11):3221-3231. doi: 10.1099/mic.0.053579-0. Epub 2011 Aug 26. Microbiology (Reading). 2011. PMID: 21873409 Free PMC article.
-
Fur activates the expression of Salmonella enterica pathogenicity island 1 by directly interacting with the hilD operator in vivo and in vitro.PLoS One. 2011 May 6;6(5):e19711. doi: 10.1371/journal.pone.0019711. PLoS One. 2011. PMID: 21573071 Free PMC article.
-
Regulation of the Bacillus subtilis fur and perR genes by PerR: not all members of the PerR regulon are peroxide inducible.J Bacteriol. 2002 Jun;184(12):3276-86. doi: 10.1128/JB.184.12.3276-3286.2002. J Bacteriol. 2002. PMID: 12029044 Free PMC article.
-
Heterogeneity in respiratory electron transfer and adaptive iron utilization in a bacterial biofilm.Nat Commun. 2019 Aug 16;10(1):3702. doi: 10.1038/s41467-019-11681-0. Nat Commun. 2019. PMID: 31420537 Free PMC article.
References
-
- Althaus E W, Outten C E, Ohlsen K E, Cao H, O’Halloran T V. The ferric uptake regulation (Fur) repressor is a zinc metalloprotein. Biochemistry. 1999;38:6559–6569. - PubMed
-
- Arnow L E. Colorimetric determination of the components of 3,4 dihydroxyphenylalanine-tyrosine mixtures. J Biol Chem. 1937;228:531–537.
-
- Bagg A, Neilands J B. Ferric uptake regulation protein acts as a repressor, employing iron (II) as a cofactor to bind the operator of an iron transport operon in Escherichia coli. Biochemistry. 1987;26:5471–5477. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

