An additional regulatory gene for actinorhodin production in Streptomyces lividans involves a LysR-type transcriptional regulator
- PMID: 10400594
- PMCID: PMC93938
- DOI: 10.1128/JB.181.14.4353-4364.1999
An additional regulatory gene for actinorhodin production in Streptomyces lividans involves a LysR-type transcriptional regulator
Abstract
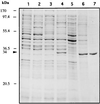
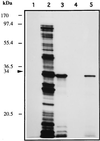
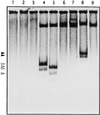
The sequence of a 4.8-kbp DNA fragment adjacent to the right-hand end of the actinorhodin biosynthetic (act) cluster downstream of actVB-orf6 from Streptomyces coelicolor A3(2) reveals six complete open reading frames, named orf7 to orf12. The deduced amino acid sequences from orf7, orf10, and orf11 show significant similarities with the following products in the databases: a putative protein from the S. coelicolor SCP3 plasmid, LysR-type transcriptional regulators, and proteins belonging to the family of short-chain dehydrogenases/reductases, respectively. The deduced product of orf8 reveals low similarities with several methyltransferases from different sources, while orf9 and orf12 products show no similarities with other known proteins. Disruptions of orf10 and orf11 genes in S. coelicolor appear to have no significant effect on the production of actinorhodin. Nevertheless, disruption or deletion of orf10 in Streptomyces lividans causes actinorhodin overproduction. The introduction of extra copies of orf10 and orf11 genes in an S. coelicolor actIII mutant restores the ability to produce actinorhodin. Transcriptional analysis and DNA footprinting indicate that Orf10 represses its own transcription and regulates orf11 transcription, expression of which might require the presence of an unknown inducer. No DNA target for Orf10 protein was found within the act cluster.
Figures

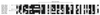






Similar articles
-
A polyketide biosynthetic gene cluster from Streptomyces antibioticus includes a LysR-type transcriptional regulator.Microbiology (Reading). 2001 Nov;147(Pt 11):3083-92. doi: 10.1099/00221287-147-11-3083. Microbiology (Reading). 2001. PMID: 11700358
-
afsB stimulates transcription of the actinorhodin biosynthetic pathway in Streptomyces coelicolor A3(2) and Streptomyces lividans.Mol Gen Genet. 1989 Jan;215(2):355-7. doi: 10.1007/BF00339742. Mol Gen Genet. 1989. PMID: 2710105
-
afsR2: a previously undetected gene encoding a 63-amino-acid protein that stimulates antibiotic production in Streptomyces lividans.Mol Microbiol. 1994 Nov;14(4):643-53. doi: 10.1111/j.1365-2958.1994.tb01303.x. Mol Microbiol. 1994. PMID: 7891553
-
1995 Colworth Prize Lecture. The regulation of antibiotic production in Streptomyces coelicolor A3(2).Microbiology (Reading). 1996 Jun;142 ( Pt 6):1335-1344. doi: 10.1099/13500872-142-6-1335. Microbiology (Reading). 1996. PMID: 8704973 Review. No abstract available.
-
Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins.Microbiology (Reading). 2008 Dec;154(Pt 12):3609-3623. doi: 10.1099/mic.0.2008/022772-0. Microbiology (Reading). 2008. PMID: 19047729 Review.
Cited by
-
Characterization of TsaR, an oxygen-sensitive LysR-type regulator for the degradation of p-toluenesulfonate in Comamonas testosteroni T-2.Appl Environ Microbiol. 2003 Apr;69(4):2298-305. doi: 10.1128/AEM.69.4.2298-2305.2003. Appl Environ Microbiol. 2003. PMID: 12676713 Free PMC article.
-
Genetically modified bacterial strains and novel bacterial artificial chromosome shuttle vectors for constructing environmental libraries and detecting heterologous natural products in multiple expression hosts.Appl Environ Microbiol. 2004 Apr;70(4):2452-63. doi: 10.1128/AEM.70.4.2452-2463.2004. Appl Environ Microbiol. 2004. PMID: 15066844 Free PMC article.
-
Biosynthesis of polyketides in heterologous hosts.Microbiol Mol Biol Rev. 2001 Mar;65(1):106-18. doi: 10.1128/MMBR.65.1.106-118.2001. Microbiol Mol Biol Rev. 2001. PMID: 11238987 Free PMC article. Review.
-
The DNA cytosine methylome revealed two methylation motifs in the upstream regions of genes related to morphological and physiological differentiation in Streptomyces coelicolor A(3)2 M145.Sci Rep. 2023 Apr 29;13(1):7038. doi: 10.1038/s41598-023-34075-1. Sci Rep. 2023. PMID: 37120673 Free PMC article.
-
The Pathway-Specific Regulator ClaR of Streptomyces clavuligerus Has a Global Effect on the Expression of Genes for Secondary Metabolism and Differentiation.Appl Environ Microbiol. 2015 Oct;81(19):6637-48. doi: 10.1128/AEM.00916-15. Epub 2015 Jul 17. Appl Environ Microbiol. 2015. PMID: 26187955 Free PMC article.
References
-
- Arrowsmith T J, Malpartida F, Sherman D H, Birch A, Hopwood D A, Robinson J A. Characterisation of actI-homologous DNA encoding polyketide synthase genes from the monensin producer Streptomyces cinnamonensis. Mol Gen Genet. 1992;234:254–264. - PubMed
-
- Bartolomé B, Jubete Y, Martínez E, de la Cruz F. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene. 1991;102:75–78. - PubMed
-
- Bibb M J, Findlay P R, Johnson M W. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene. 1984;30:157–166. - PubMed
-
- Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical

