Isolation and characterization of mutations in the Escherichia coli regulatory protein XapR
- PMID: 10400599
- PMCID: PMC93943
- DOI: 10.1128/JB.181.14.4397-4403.1999
Isolation and characterization of mutations in the Escherichia coli regulatory protein XapR
Abstract
In this work, the LysR-type protein XapR has been subjected to a mutational analysis. XapR regulates the expression of xanthosine phosphorylase (XapA), a purine nucleoside phosphorylase in Escherichia coli. In the wild type, full expression of XapA requires both a functional XapR protein and the inducer xanthosine. Here we show that deoxyinosine can also function as an inducer in the wild type, although not to the same extent as xanthosine. We have isolated and characterized in detail the mutants that can be induced by other nucleosides as well as xanthosine. Sequencing of the mutants has revealed that two regions in XapR are important for correct interactions between the inducer and XapR. One region is defined by amino acids 104 and 132, and the other region, containing most of the isolated mutations, is found between amino acids 203 and 210. These regions, when modelled into the three-dimensional structure of CysB from Klebsiella aerogenes, are placed close together and are most probably directly involved in binding the inducer xanthosine.
Figures
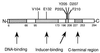
Similar articles
-
Xanthosine utilization in Salmonella enterica serovar Typhimurium is recovered by a single aspartate-to-glycine substitution in xanthosine phosphorylase.J Bacteriol. 2006 Jun;188(11):4153-7. doi: 10.1128/JB.01926-05. J Bacteriol. 2006. PMID: 16707709 Free PMC article.
-
Identification and characterization of genes (xapA, xapB, and xapR) involved in xanthosine catabolism in Escherichia coli.J Bacteriol. 1995 Oct;177(19):5506-16. doi: 10.1128/jb.177.19.5506-5516.1995. J Bacteriol. 1995. PMID: 7559336 Free PMC article.
-
[Regulatory mutants for the synthesis of a 2d purine nucleoside phosphorylase in Escherichia coli K-12. I. Synthesis inducers and the substrate specificity of purine nucleoside phosphorylase in pndR mutants].Genetika. 1984 Sep;20(9):1463-71. Genetika. 1984. PMID: 6437906 Russian.
-
[Purine nucleoside phosphorylase (PNP)].Nihon Rinsho. 2003 Jan;61 Suppl 1:85-90. Nihon Rinsho. 2003. PMID: 12629696 Review. Japanese. No abstract available.
-
The molecular basis for positive regulation of cys promoters in Salmonella typhimurium and Escherichia coli.Mol Microbiol. 1992 Oct;6(19):2747-53. doi: 10.1111/j.1365-2958.1992.tb01453.x. Mol Microbiol. 1992. PMID: 1435253 Review.
Cited by
-
Isolation and characterization of mutant Sinorhizobium meliloti NodD1 proteins with altered responses to luteolin.J Bacteriol. 2013 Aug;195(16):3714-23. doi: 10.1128/JB.00309-13. Epub 2013 Jun 14. J Bacteriol. 2013. PMID: 23772067 Free PMC article.
-
Regulation of expression of the tricarballylate utilization operon (tcuABC) of Salmonella enterica.Res Microbiol. 2009 Apr;160(3):179-86. doi: 10.1016/j.resmic.2009.01.001. Epub 2009 Jan 21. Res Microbiol. 2009. PMID: 19284970 Free PMC article.
-
Xanthosine utilization in Salmonella enterica serovar Typhimurium is recovered by a single aspartate-to-glycine substitution in xanthosine phosphorylase.J Bacteriol. 2006 Jun;188(11):4153-7. doi: 10.1128/JB.01926-05. J Bacteriol. 2006. PMID: 16707709 Free PMC article.
-
The structure of CrgA from Neisseria meningitidis reveals a new octameric assembly state for LysR transcriptional regulators.Nucleic Acids Res. 2009 Aug;37(14):4545-58. doi: 10.1093/nar/gkp445. Epub 2009 May 27. Nucleic Acids Res. 2009. PMID: 19474343 Free PMC article.
-
CbbR, the Master Regulator for Microbial Carbon Dioxide Fixation.J Bacteriol. 2015 Nov;197(22):3488-98. doi: 10.1128/JB.00442-15. Epub 2015 Aug 31. J Bacteriol. 2015. PMID: 26324454 Free PMC article. Review.
References
-
- Bartowsky E, Normark S. Purification and mutant analysis of Citrobacter freundii AmpR, the regulator for chromosomal AmpC beta-lactamase. Mol Microbiol. 1991;5:1715–1725. - PubMed
-
- Burn J E, Hamilton W D, Wootton J C, Johnston A W B. Single and multiple mutations affecting properties of the regulatory gene nodD of Rhizobium. Mol Microbiol. 1989;3:1567–1577. - PubMed
-
- Buxton R S, Hammer-Jespersen K H, Valentin-Hansen P. A second purine nucleoside phosphorylase in Escherichia coli K-12. I. Xanthosine phosphorylase regulatory mutants isolated as secondary-site revertants of a deoD mutant. Mol Gen Genet. 1980;179:331–340. - PubMed
-
- Cebolla A, Sousa C, de Lorenzo V. Effector specificity mutants of the transcriptional activator NahR of naphthalene degrading Pseudomonas define protein sites involved in binding of aromatic inducers. J Biol Chem. 1997;272:3986–3992. - PubMed
-
- Clark D J, Maaløe O. DNA replication and the division cycle of Escherichia coli. J Mol Biol. 1967;23:99–112.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

