The gag domains required for avian retroviral RNA encapsidation determined by using two independent assays
- PMID: 10400719
- PMCID: PMC112706
- DOI: 10.1128/JVI.73.8.6282-6292.1999
The gag domains required for avian retroviral RNA encapsidation determined by using two independent assays
Abstract
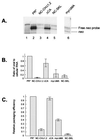
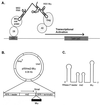
The Rous sarcoma virus (RSV) Gag precursor polyprotein is the only viral protein which is necessary for specific packaging of genomic RNA. To map domains within Gag which are important for packaging, we constructed a series of Gag mutations in conjunction with a protease (PR) active-site point mutation in a full-length viral construct. We found that deletion of either the matrix (MA), the capsid (CA), or the protease (PR) domain did not abrogate packaging, although the MA domain is likely to be required for proper assembly. A previously characterized deletion of both Cys-His motifs in RSV nucleocapsid protein (NC) reduced both the efficiency of particle release and specific RNA packaging by 6- to 10-fold, consistent with previous observations that the NC Cys-His motifs played a role in assembly and RNA packaging. Most strikingly, when amino acid changes at Arg 549 and 551 immediately downstream of the distal NC Cys-His box were made, RNA packaging was reduced by more than 25-fold with no defect in particle release, demonstrating the importance of this basic amino acid region in packaging. We also used the yeast three-hybrid system to study avian retroviral RNA-Gag interactions. Using this assay, we found that the interactions of the minimal packaging region (Mpsi) with Gag are of high affinity and specificity. Using a number of Mpsi and Gag mutants, we have found a clear correlation between a reporter gene activation in a yeast three-hybrid binding system and an in vivo packaging assay. Our results showed that the binding assay provides a rapid genetic assay of both RNA and protein components for specific encapsidation.
Figures






References
-
- Allen P, Collins B, Brown D, Hostomsky Z, Gold L. A specific RNA structural motif mediates high affinity binding by the HIV-1 nucleocapsid protein (NCp7) Virology. 1996;225:306–315. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

