A lysine-to-arginine change found in natural alleles of the human T-cell lymphotropic/leukemia virus type 1 p12(I) protein greatly influences its stability
- PMID: 10400740
- PMCID: PMC112727
- DOI: 10.1128/JVI.73.8.6460-6467.1999
A lysine-to-arginine change found in natural alleles of the human T-cell lymphotropic/leukemia virus type 1 p12(I) protein greatly influences its stability
Abstract
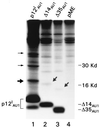
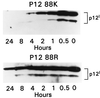
The HTLV-1 singly spliced open reading frame I protein, p12(I), is highly unstable and appears to be necessary for persistent infection in rabbits. Here we demonstrate that p12(I) forms dimers through two putative leucine zipper domains and that its stability is augmented by specific proteasome inhibitors. p12(I) is ubiquitylated, and mutations of its unique carboxy-terminus lysine residue to an arginine greatly enhance its stability. Interestingly, analysis of 53 independent HTLV-1 strains revealed that the natural p12(I) alleles found in ex vivo samples of tropical spastic paraparesis-HTLV-1-associated myelopathy patients contain a Lys at position 88 in some cases, whereas arginine is consistently found at position 88 in HTLV-1 strains from all adult T-cell leukemia-lymphoma (ATLL) cases and healthy carriers studied. This apparent segregation of different alleles in tropical spastic paraparesis-HTLV-associated myelopathy and ATLL or healthy carriers may be relevant in vivo, since p12(I) binds the interleukin-2 receptor beta and gammac chains, raising the possibility that the two natural alleles might affect differently the regulation of these molecules.
Figures






Similar articles
-
Frequency of p12K and p12R alleles of HTLV Type 1 in HAM/TSP patients and in asymptomatic HTLV type 1 carriers.AIDS Res Hum Retroviruses. 2002 Sep 1;18(13):899-902. doi: 10.1089/088922202760265560. AIDS Res Hum Retroviruses. 2002. PMID: 12230932
-
Human T cell lymphotropic virus type I (HTLV-I) p12I is dispensable for HTLV-I transmission and maintenance of infection in vivo.AIDS Res Hum Retroviruses. 2004 Oct;20(10):1092-9. doi: 10.1089/aid.2004.20.1092. AIDS Res Hum Retroviruses. 2004. PMID: 15585100
-
Simian T cell leukemia virus type I from naturally infected feral monkeys from central and west Africa encodes a 91-amino acid p12 (ORF-I) protein as opposed to a 99-amino acid protein encoded by HTLV type I from humans.AIDS Res Hum Retroviruses. 1997 Mar 20;13(5):425-32. doi: 10.1089/aid.1997.13.425. AIDS Res Hum Retroviruses. 1997. PMID: 9075484
-
[HTLV-1: Recent topics in epidemiologic, basic and clinical research].Uirusu. 2013;63(2):165-74. doi: 10.2222/jsv.63.165. Uirusu. 2013. PMID: 25366051 Review. Japanese.
-
Human T-lymphotropic virus type 1 non-structural proteins: Requirements for latent infection.Cancer Sci. 2013 Aug;104(8):983-8. doi: 10.1111/cas.12190. Epub 2013 Jun 17. Cancer Sci. 2013. PMID: 23651172 Free PMC article. Review.
Cited by
-
Lessons from viral manipulation of protein disposal pathways.J Clin Invest. 2002 Oct;110(7):875-9. doi: 10.1172/JCI16831. J Clin Invest. 2002. PMID: 12370262 Free PMC article. Review. No abstract available.
-
Non-Structural Proteins from Human T-cell Leukemia Virus Type 1 in Cellular Membranes-Mechanisms for Viral Survivability and Proliferation.Int J Mol Sci. 2018 Nov 8;19(11):3508. doi: 10.3390/ijms19113508. Int J Mol Sci. 2018. PMID: 30413005 Free PMC article. Review.
-
Role of accessory proteins of HTLV-1 in viral replication, T cell activation, and cellular gene expression.Front Biosci. 2004 Sep 1;9:2556-76. doi: 10.2741/1417. Front Biosci. 2004. PMID: 15358581 Free PMC article. Review.
-
In vivo genetic mutations define predominant functions of the human T-cell leukemia/lymphoma virus p12I protein.Blood. 2009 Apr 16;113(16):3726-34. doi: 10.1182/blood-2008-04-146928. Epub 2008 Sep 12. Blood. 2009. PMID: 18791162 Free PMC article.
-
Hijacking the T-cell communication network by the human T-cell leukemia/lymphoma virus type 1 (HTLV-1) p12 and p8 proteins.Mol Aspects Med. 2010 Oct;31(5):333-43. doi: 10.1016/j.mam.2010.07.001. Epub 2010 Jul 29. Mol Aspects Med. 2010. PMID: 20673780 Free PMC article. Review.
References
-
- Berneman Z N, Gartenhaus R B, Reitz M S, Jr, Blattner W A, Manns A, Hanchard B, Ikehara O, Gallo R C, Klotman M E. Expression of alternatively spliced human T-lymphotropic virus type 1 (HTLV-I) pX mRNA in infected cell lines and in primary uncultured cells from patients with adult T-cell leukemia/lymphoma and healthy carriers. Proc Natl Acad Sci USA. 1992;89:3005–3009. - PMC - PubMed
-
- Bonifacino J S. Reversal of fortune for nascent proteins. Nature. 1996;384:405–406. - PubMed
-
- Collins N D, Newbound G C, Albrecht B, Beard J L, Ratner L, Lairmore M D. Selective ablation of human T-cell lymphotropic virus type 1 p12I reduces viral infectivity in vivo. Blood. 1998;91:4701–4707. - PubMed
-
- Derse D, Mikovits J, Ruscetti F. X-I and X-II open reading frames of HTLV-I are not required for virus replication or for immortalization of primary T-cells in vitro. Virology. 1997;237:123–128. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

