Nonproductive human immunodeficiency virus type 1 infection in nucleoside-treated G0 lymphocytes
- PMID: 10400748
- PMCID: PMC112735
- DOI: 10.1128/JVI.73.8.6526-6532.1999
Nonproductive human immunodeficiency virus type 1 infection in nucleoside-treated G0 lymphocytes
Abstract
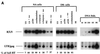
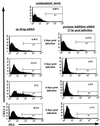
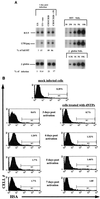
Productive infection by human immunodeficiency virus type 1 (HIV-1) requires the activation of target cells. Infection of quiescent peripheral CD4 lymphocytes by HIV-1 results in incomplete, labile, reverse transcripts. We have previously identified G1b as the cell cycle stage required for the optimal completion of the reverse transcription process in T lymphocytes. However, the mechanism(s) involved in the blockage of reverse transcription remains undefined. In this study we investigated whether nucleotide levels influence viral reverse transcription in G0 cells. For this purpose the role of the enzyme ribonucleotide reductase was bypassed, by adding exogenous deoxyribonucleosides to highly purified T cells in the G0 or the G1a phase of the cell cycle. Our data showed a significant increase in the efficiency of the reverse transcription process following the addition of the deoxyribonucleosides. To define the stability and functionality of these full reverse transcripts, we used an HIV-1 reporter virus that expresses the murine heat-stable antigen on the surfaces of infected cells. Following activation of infected quiescent cells treated with exogenous nucleosides, no increased rescue of productive infection was seen. Thus, in addition to failure to complete reverse transcription, there was an additional nonreversible blockage of productive infection in quiescent T cells. These experiments have important relevance in the gene therapy arena, in terms of improving the ability of lentivirus vectors to enter metabolically inactive cells, such as hematopoietic stem cells.
Figures


References
-
- Cohen A, Barankiewicz J, Lederman H M, Gelfand E W. Purine and pyrimidine metabolism in human T lymphocytes. Regulation of deoxyribonucleotide metabolism. J Biol Chem. 1983;258:12334–12340. - PubMed
-
- Eriksson S, Thelander L, Akerman M. Allosteric regulation of calf thymus ribonucleoside diphosphate reductase. Biochemistry. 1979;18:2948–2952. - PubMed
-
- Gao W Y, Agbaria R, Driscoll J S, Mitsuya H. Divergent anti-human immunodeficiency virus activity and anabolic phosphorylation of 2′,3′-dideoxynucleoside analogs in resting and activated human cells. J Biol Chem. 1994;269:12633–12638. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

