Mutagenesis of CXCR4 identifies important domains for human immunodeficiency virus type 1 X4 isolate envelope-mediated membrane fusion and virus entry and reveals cryptic coreceptor activity for R5 isolates
- PMID: 10400757
- PMCID: PMC112744
- DOI: 10.1128/JVI.73.8.6598-6609.1999
Mutagenesis of CXCR4 identifies important domains for human immunodeficiency virus type 1 X4 isolate envelope-mediated membrane fusion and virus entry and reveals cryptic coreceptor activity for R5 isolates
Abstract
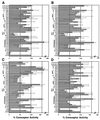
CXCR4 is a chemokine receptor and a coreceptor for T-cell-line-tropic (X4) and dual-tropic (R5X4) human immunodeficiency virus type 1 (HIV-1) isolates. Cells coexpressing CXCR4 and CD4 will fuse with appropriate HIV-1 envelope glycoprotein (Env)-expressing cells. The delineation of the critical regions involved in the interactions within the Env-CD4-coreceptor complex are presently under intensive investigation, and the use of chimeras of coreceptor molecules has provided valuable information. To define these regions in greater detail, we have employed a strategy involving alanine-scanning mutagenesis of the extracellular domains of CXCR4 coupled with a highly sensitive reporter gene assay for HIV-1 Env-mediated membrane fusion. Using a panel of 41 different CXCR4 mutants, we have identified several charged residues that appear important for coreceptor activity for X4 Envs; the mutations E15A (in which the glutamic acid residue at position 15 is replaced by alanine) and E32A in the N terminus, D97A in extracellular loop 1 (ecl-1), and R188A in ecl-2 impaired coreceptor activity for X4 and R5X4 Envs. In addition, substitution of alanine for any of the four extracellular cysteines alone resulted in conformational changes of various degrees, while mutants with paired cysteine deletions partially retained their structure. Our data support the notion that all four cysteines are involved in disulfide bond formation. We have also identified substitutions which greatly enhance or convert CXCR4's coreceptor activity to support R5 Env-mediated fusion (N11A, R30A, D187A, and D193A), and together our data suggest the presence of conserved extracellular elements, common to both CXCR4 and CCR5, involved in their coreceptor activities. These data will help us to better detail the CXCR4 structural requirements exhibited by different HIV-1 strains and will direct further mutagenesis efforts aimed at better defining the domains in CXCR4 involved in the HIV-1 Env-mediated fusion process.
Figures






References
-
- Atchison R E, Gosling J, Monteclaro F S, Franci C, Digilio L, Charo I F, Goldsmith M A. Multiple extracellular elements of CCR5 and HIV-1 entry: dissociation from response to chemokines. Science. 1996;274:1924–1926. - PubMed
-
- Berger E A, Doms R W, Fenyo E M, Korber B T, Littman D R, Moore J P, Sattentau Q J, Schuitemaker H, Sodroski J, Weiss R A. A new classification for HIV-1. Nature. 1998;391:240. . (Letter.) - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

