Coupled integration of human immunodeficiency virus type 1 cDNA ends by purified integrase in vitro: stimulation by the viral nucleocapsid protein
- PMID: 10400764
- PMCID: PMC112751
- DOI: 10.1128/JVI.73.8.6670-6679.1999
Coupled integration of human immunodeficiency virus type 1 cDNA ends by purified integrase in vitro: stimulation by the viral nucleocapsid protein
Abstract
Integration of retroviral cDNA involves coupled joining of the two ends of the viral genome at precisely spaced positions in the host cell DNA. Correct coupled joining is essential for viral replication, as shown, for example, by the finding that viral mutants defective in coupled joining are defective in integration and replication. To date, reactions with purified human immunodeficiency virus type 1 (HIV-1) integrase protein in vitro have supported mainly uncoupled joining of single cDNA ends. We have analyzed an activity stimulating coupled joining present in HIV-1 virions, which led to the finding that the HIV-1 nucleocapsid (NC) protein can stimulate coupled joining more than 1,000-fold under some conditions. The requirements for stimulating coupled joining were investigated in assays with mutant NC proteins, revealing that mutations in the zinc finger domains can influence stimulation of integration. These findings (i) provide a means for assembling more authentic integrase complexes for mechanistic studies, (ii) reveal a new activity of NC protein in vitro, (iii) indicate a possible role for NC in vivo, and (iv) provide a possible method for identifying a new class of inhibitors that disrupt coupled joining.
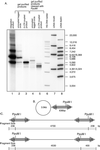
Figures






References
-
- Allen P, Worland S, Gold L. Isolation of high-affinity RNA ligands to HIV-1 integrase from a random pool. Virology. 1995;209:327–336. - PubMed
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons, Inc.; 1987.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

