Structure of adenovirus complexed with its internalization receptor, alphavbeta5 integrin
- PMID: 10400774
- PMCID: PMC112761
- DOI: 10.1128/JVI.73.8.6759-6768.1999
Structure of adenovirus complexed with its internalization receptor, alphavbeta5 integrin
Abstract
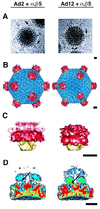
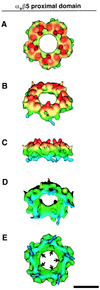
The three-dimensional structure of soluble recombinant integrin alphavbeta5 bound to human adenovirus types 2 and 12 (Ad2 and -12) has been determined at approximately 21-A resolution by cryoelectron microscopy (cryo-EM). The alphavbeta5 integrin is known to promote Ad cell entry. Cryo-EM has shown that the integrin-binding RGD (Arg-Gly-Asp) protrusion of the Ad2 penton base protein is highly mobile (P. L. Stewart, C. Y. Chiu, S. Huang, T. Muir, Y. Zhao, B. Chait, P. Mathias, and G. R. Nemerow, EMBO J. 16:1189-1198, 1997). Sequence analysis indicated that the Ad12 RGD surface loop is shorter than that of Ad2 and probably less flexible, hence more suitable for structural characterization of the Ad-integrin complex. The cryo-EM structures of the two virus-receptor complexes revealed a ring of integrin density above the penton base of each virus serotype. As expected, the integrin density in the Ad2 complex was diffuse while that in the Ad12 complex was better defined. The integrin consists of two discrete subdomains, a globular domain with an RGD-binding cleft approximately 20 A in diameter and a distal domain with extended, flexible tails. Kinetic analysis of Ad2 interactions with alphavbeta5 indicated approximately 4.2 integrin molecules bound per penton base at close to saturation. These results suggest that the precise spatial arrangement of five RGD protrusions on the penton base promotes integrin clustering and the signaling events required for virus internalization.
Figures






Similar articles
-
Vitronectin receptor antibodies inhibit infection of HeLa and A549 cells by adenovirus type 12 but not by adenovirus type 2.J Virol. 1994 Sep;68(9):5925-32. doi: 10.1128/JVI.68.9.5925-5932.1994. J Virol. 1994. PMID: 7520097 Free PMC article.
-
Cryo-electron microscopy structure of an adenovirus-integrin complex indicates conformational changes in both penton base and integrin.J Virol. 2009 Nov;83(22):11491-501. doi: 10.1128/JVI.01214-09. Epub 2009 Sep 2. J Virol. 2009. PMID: 19726496 Free PMC article.
-
Comparative analysis of adenovirus fiber-cell interaction: adenovirus type 2 (Ad2) and Ad9 utilize the same cellular fiber receptor but use different binding strategies for attachment.J Virol. 1996 Nov;70(11):7614-21. doi: 10.1128/JVI.70.11.7614-7621.1996. J Virol. 1996. PMID: 8892881 Free PMC article.
-
Cell receptors involved in adenovirus entry.Virology. 2000 Aug 15;274(1):1-4. doi: 10.1006/viro.2000.0468. Virology. 2000. PMID: 10936081 Review. No abstract available.
-
Integrin-Targeting Strategies for Adenovirus Gene Therapy.Viruses. 2024 May 13;16(5):770. doi: 10.3390/v16050770. Viruses. 2024. PMID: 38793651 Free PMC article. Review.
Cited by
-
Three-dimensional model of the human platelet integrin alpha IIbbeta 3 based on electron cryomicroscopy and x-ray crystallography.Proc Natl Acad Sci U S A. 2002 Oct 29;99(22):14059-64. doi: 10.1073/pnas.212498199. Epub 2002 Oct 18. Proc Natl Acad Sci U S A. 2002. PMID: 12388784 Free PMC article.
-
Structural analysis of a fiber-pseudotyped adenovirus with ocular tropism suggests differential modes of cell receptor interactions.J Virol. 2001 Jun;75(11):5375-80. doi: 10.1128/JVI.75.11.5375-5380.2001. J Virol. 2001. PMID: 11333920 Free PMC article.
-
Engineering clustered ligand binding into nonviral vectors: alphavbeta3 targeting as an example.Mol Ther. 2009 May;17(5):828-36. doi: 10.1038/mt.2009.11. Epub 2009 Feb 24. Mol Ther. 2009. PMID: 19240693 Free PMC article.
-
Current advances and future challenges in Adenoviral vector biology and targeting.Curr Gene Ther. 2007 Jun;7(3):189-204. doi: 10.2174/156652307780859062. Curr Gene Ther. 2007. PMID: 17584037 Free PMC article. Review.
-
Integrin and defensin modulate the mechanical properties of adenovirus.J Virol. 2013 Mar;87(5):2756-66. doi: 10.1128/JVI.02516-12. Epub 2012 Dec 26. J Virol. 2013. PMID: 23269786 Free PMC article.
References
-
- Acharya R, Fry E, Stuart D, Fox G, Rowlands D, Brown F. The three-dimensional structure of foot-and-mouth disease virus at 2.9 Å resolution. Nature. 1989;337:709–716. - PubMed
-
- Adler M, Lazarus R A, Dennis M S, Wagner G. Solution structure of kistrin, a potent platelet aggregation inhibitor and GP IIb-IIIa antagonist. Science. 1991;253:445–448. - PubMed
-
- Adrian M, Dubochet J, Lepault J, McDowall A W. Cryo-electron microscopy of viruses. Nature. 1984;308:32–36. - PubMed
-
- Akke M, Liu J, Cavanagh J, Erickson H P, Palmer A G I. Pervasive conformational fluctuations on microsecond time scales in a fibronectin type III domain. Nat Struct Biol. 1998;5:55–59. - PubMed
-
- Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

