Association with the cellular export receptor CRM 1 mediates function and intracellular localization of Epstein-Barr virus SM protein, a regulator of gene expression
- PMID: 10400785
- PMCID: PMC112772
- DOI: 10.1128/JVI.73.8.6872-6881.1999
Association with the cellular export receptor CRM 1 mediates function and intracellular localization of Epstein-Barr virus SM protein, a regulator of gene expression
Abstract
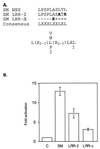
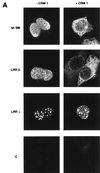
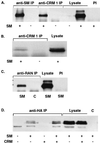
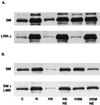
Splicing and posttranscriptional processing of eukaryotic gene transcripts are linked to their nuclear export and cytoplasmic expression. Unspliced pre-mRNAs and intronless transcripts are thus inherently poorly expressed. Nevertheless, human and animal viruses encode essential genes as single open reading frames or in the intervening sequences of other genes. Many retroviruses have evolved mechanisms to facilitate nuclear export of their unspliced mRNAs. For example, the human immunodeficiency virus RNA-binding protein Rev associates with the soluble cellular export receptor CRM 1 (exportin 1), which mediates nucleocytoplasmic translocation of Rev-HIV RNA complexes through the nuclear pore. The transforming human herpesvirus Epstein-Barr virus (EBV) expresses a nuclear protein, SM, early in its lytic cycle; SM binds RNA and posttranscriptionally activates expression of certain intronless lytic EBV genes. Here we show that both the trans-activation function and cytoplasmic translocation of SM are dependent on association with CRM 1 in vivo. SM is also shown to be associated in vivo with other components of the CRM 1 export pathway, including the small GTPase Ran and the nucleoporin CAN/Nup214. SM is shown to be present in the cytoplasm, nucleoplasm, and nuclear envelope of transfected cells. Mutation of a leucine-rich region (LRR) of SM inhibited CRM 1-mediated cytoplasmic translocation and SM activity, as did leptomycin B, an inhibitor of CRM 1 complex formation. Surprisingly, however, leptomycin B treatment and mutation of the LRR both led to SM becoming more tightly attached to intranuclear structures. These findings suggest a model in which SM is not merely a soluble carrier protein for RNA but rather is bound directly to intranuclear proteins, possibly including the nuclear pore complex.
Figures




References
-
- Agard D A, Hiraoka Y, Shaw P, Sedat J W. Fluorescence microscopy in three dimensions. Methods Cell Biol. 1989;30:353–377. - PubMed
-
- Baer R, Bankier A T, Biggin M D. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984;310:207–211. - PubMed
-
- Bogerd H P, Echarri A, Ross T M, Cullen B R. Inhibition of human immunodeficiency virus Rev and human T-cell leukemia virus Rex function, but not Mason-Pfizer monkey virus constitutive transport element activity, by a mutant human nucleoporin targeted to Crm1. J Virol. 1998;72:8627–8635. - PMC - PubMed
-
- Callis J, Fromm M, Walbot V. Introns increase gene expression in cultured maize cells. Genes Dev. 1987;1:1183–1200. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

