Three-dimensional structure of Aleutian mink disease parvovirus: implications for disease pathogenicity
- PMID: 10400786
- PMCID: PMC112773
- DOI: 10.1128/JVI.73.8.6882-6891.1999
Three-dimensional structure of Aleutian mink disease parvovirus: implications for disease pathogenicity
Abstract
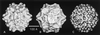
The three-dimensional structure of expressed VP2 capsids of Aleutian mink disease parvovirus strain G (ADVG-VP2) has been determined to 22 A resolution by cryo-electron microscopy and image reconstruction techniques. A structure-based sequence alignment of the VP2 capsid protein of canine parvovirus (CPV) provided a means to construct an atomic model of the ADVG-VP2 capsid. The ADVG-VP2 reconstruction reveals a capsid structure with a mean external radius of 128 A and several surface features similar to those found in human parvovirus B19 (B19), CPV, feline panleukopenia virus (FPV), and minute virus of mice (MVM). Dimple-like depressions occur at the icosahedral twofold axes, canyon-like regions encircle the fivefold axes, and spike-like protrusions decorate the threefold axes. These spikes are not present in B19, and they are more prominent in ADV compared to the other parvoviruses owing to the presence of loop insertions which create mounds near the threefold axes. Cylindrical channels along the fivefold axes of CPV, FPV, and MVM, which are surrounded by five symmetry-related beta-ribbons, are closed in ADVG-VP2 and B19. Immunoreactive peptides made from segments of the ADVG-VP2 capsid protein map to residues in the mound structures. In vitro tissue tropism and in vivo pathogenic properties of ADV map to residues at the threefold axes and to the wall of the dimples.
Figures


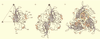



Similar articles
-
Identification of aleutian mink disease parvovirus capsid sequences mediating antibody-dependent enhancement of infection, virus neutralization, and immune complex formation.J Virol. 2001 Nov;75(22):11116-27. doi: 10.1128/JVI.75.22.11116-11127.2001. J Virol. 2001. PMID: 11602751 Free PMC article.
-
The relationship between capsid protein (VP2) sequence and pathogenicity of Aleutian mink disease parvovirus (ADV): a possible role for raccoons in the transmission of ADV infections.J Virol. 1996 Feb;70(2):852-61. doi: 10.1128/JVI.70.2.852-861.1996. J Virol. 1996. PMID: 8551624 Free PMC article.
-
Replication of Aleutian mink disease parvovirus in vivo is influenced by residues in the VP2 protein.J Virol. 1999 Oct;73(10):8713-9. doi: 10.1128/JVI.73.10.8713-8719.1999. J Virol. 1999. PMID: 10482625 Free PMC article.
-
Pathogenesis of aleutian mink disease parvovirus and similarities to b19 infection.J Vet Med B Infect Dis Vet Public Health. 2005 Sep-Oct;52(7-8):331-4. doi: 10.1111/j.1439-0450.2005.00864.x. J Vet Med B Infect Dis Vet Public Health. 2005. PMID: 16316395 Review.
-
Molecular and structural basis of the evolution of parvovirus tropism.Acta Vet Hung. 1999;47(3):379-94. doi: 10.1556/AVet.47.1999.3.11. Acta Vet Hung. 1999. PMID: 10497831 Review.
Cited by
-
Minute virus of mice, a parvovirus, in complex with the Fab fragment of a neutralizing monoclonal antibody.J Virol. 2007 Sep;81(18):9851-8. doi: 10.1128/JVI.00775-07. Epub 2007 Jul 11. J Virol. 2007. PMID: 17626084 Free PMC article.
-
Surface-exposed adeno-associated virus Vp1-NLS capsid fusion protein rescues infectivity of noninfectious wild-type Vp2/Vp3 and Vp3-only capsids but not that of fivefold pore mutant virions.J Virol. 2007 Aug;81(15):7833-43. doi: 10.1128/JVI.00580-07. Epub 2007 May 16. J Virol. 2007. PMID: 17507473 Free PMC article.
-
Identification of aleutian mink disease parvovirus capsid sequences mediating antibody-dependent enhancement of infection, virus neutralization, and immune complex formation.J Virol. 2001 Nov;75(22):11116-27. doi: 10.1128/JVI.75.22.11116-11127.2001. J Virol. 2001. PMID: 11602751 Free PMC article.
-
Molecular epidemiology of Aleutian disease virus in free-ranging domestic, hybrid, and wild mink.Evol Appl. 2012 Jun;5(4):330-40. doi: 10.1111/j.1752-4571.2011.00224.x. Epub 2012 Apr 12. Evol Appl. 2012. PMID: 25568054 Free PMC article.
-
Adding the third dimension to virus life cycles: three-dimensional reconstruction of icosahedral viruses from cryo-electron micrographs.Microbiol Mol Biol Rev. 1999 Dec;63(4):862-922, table of contents. doi: 10.1128/MMBR.63.4.862-922.1999. Microbiol Mol Biol Rev. 1999. PMID: 10585969 Free PMC article. Review.
References
-
- Adrian M, Dubochet J, Lepault J, McDowall A W. Cryo-electron microscopy of viruses. Nature. 1984;308:32–36. - PubMed
-
- Agbandje-McKenna M, Llamas-Saiz A L, Wang F, Tattersall P, Rossmann M G. Functional implications of the structure of the murine parvovirus, minute virus of mice. Structure. 1998;6:1369–1381. - PubMed
-
- Agbandje M, Kajigaya S, McKenna R, Young N S, Rossmann M G. The structure of human parvovirus B19 at 8 Å resolution. Virology. 1994;203:106–115. - PubMed
-
- Agbandje M, McKenna R, Rossmann M G, Strassheim M L, Parish C R. Structure determination of feline panleukopenia virus empty particles. Proteins. 1993;16:155–171. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

