Specific interaction between the hepatitis C virus NS5B RNA polymerase and the 3' end of the viral RNA
- PMID: 10400807
- PMCID: PMC112794
- DOI: 10.1128/JVI.73.8.7044-7049.1999
Specific interaction between the hepatitis C virus NS5B RNA polymerase and the 3' end of the viral RNA
Abstract
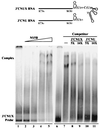
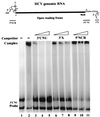
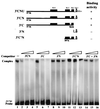
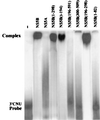
Hepatitis C virus (HCV) NS5B protein is the viral RNA-dependent RNA polymerase capable of directing RNA synthesis. In this study, an electrophoretic mobility shift assay demonstrated the interaction between a partially purified recombinant NS5B protein and a 3' viral genomic RNA with or without the conserved 98-nucleotide tail. The NS5B-RNA complexes were specifically competed away by the unlabeled homologous RNA but not by the viral 5' noncoding region and very poorly by the 3' conserved 98-nucleotide tail. A 3' coding region with conserved stem-loop structures rather than the 3' noncoding region of the HCV genome is critical for the specific binding of NS5B. Nevertheless, no direct interaction between the 3' coding region and the HCV NS5A protein was detected. Furthermore, two independent RNA-binding domains (RBDs) of NS5B were identified, RBD1, from amino acid residues 83 to 194, and RBD2, from residues 196 to 298. Interestingly, the conserved motifs of RNA-dependent RNA polymerase for putative RNA binding (220-DxxxxD-225) and template/primer position (282-S/TGxxxTxxxNS/T-292) are present in the RBD2. Nevertheless, the RNA-binding activity of RBD2 was abolished when it was linked to the carboxy-terminal half of the NS5B. These results provide some clues to understanding the initiation of HCV replication.
Figures


Similar articles
-
Template requirements for de novo RNA synthesis by hepatitis C virus nonstructural protein 5B polymerase on the viral X RNA.J Virol. 2002 Jul;76(14):6944-56. doi: 10.1128/jvi.76.14.6944-6956.2002. J Virol. 2002. PMID: 12072495 Free PMC article.
-
Enzymatic characterization of the full-length and C-terminally truncated hepatitis C virus RNA polymerases: function of the last 21 amino acids of the C terminus in template binding and RNA synthesis.Biochemistry. 2004 Aug 17;43(32):10579-91. doi: 10.1021/bi049773g. Biochemistry. 2004. PMID: 15301555
-
Template requirement and initiation site selection by hepatitis C virus polymerase on a minimal viral RNA template.J Biol Chem. 2000 Jun 9;275(23):17710-7. doi: 10.1074/jbc.M908781199. J Biol Chem. 2000. PMID: 10749880
-
Hepatitis C virus RNA-dependent RNA polymerase (NS5B polymerase).Curr Top Microbiol Immunol. 2000;242:225-60. doi: 10.1007/978-3-642-59605-6_11. Curr Top Microbiol Immunol. 2000. PMID: 10592663 Review. No abstract available.
-
Using the Hepatitis C Virus RNA-Dependent RNA Polymerase as a Model to Understand Viral Polymerase Structure, Function and Dynamics.Viruses. 2015 Jul 17;7(7):3974-94. doi: 10.3390/v7072808. Viruses. 2015. PMID: 26193306 Free PMC article. Review.
Cited by
-
The Role of the RNA-RNA Interactome in the Hepatitis C Virus Life Cycle.Int J Mol Sci. 2020 Feb 21;21(4):1479. doi: 10.3390/ijms21041479. Int J Mol Sci. 2020. PMID: 32098260 Free PMC article. Review.
-
Template requirements for de novo RNA synthesis by hepatitis C virus nonstructural protein 5B polymerase on the viral X RNA.J Virol. 2002 Jul;76(14):6944-56. doi: 10.1128/jvi.76.14.6944-6956.2002. J Virol. 2002. PMID: 12072495 Free PMC article.
-
Structural characterization of the highly conserved 98-base sequence at the 3' end of HCV RNA genome and the complementary sequence located at the 5' end of the replicative viral strand.Nucleic Acids Res. 2005 Jan 28;33(2):693-703. doi: 10.1093/nar/gki218. Print 2005. Nucleic Acids Res. 2005. PMID: 15681619 Free PMC article.
-
RNA binding protein 24 regulates the translation and replication of hepatitis C virus.Protein Cell. 2018 Nov;9(11):930-944. doi: 10.1007/s13238-018-0507-x. Epub 2018 Jan 30. Protein Cell. 2018. PMID: 29380205 Free PMC article.
-
Let-7b is a novel regulator of hepatitis C virus replication.Cell Mol Life Sci. 2012 Aug;69(15):2621-33. doi: 10.1007/s00018-012-0940-6. Epub 2012 Mar 6. Cell Mol Life Sci. 2012. PMID: 22391672 Free PMC article.
References
-
- Al R H, Xie Y, Wang Y, Hagedorn C H. Expression of recombinant hepatitis C virus non-structural protein 5B in Escherichia coli. Virus Res. 1998;53:141–149. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

