Human immunodeficiency virus type 1 tat protein activates transcription factor NF-kappaB through the cellular interferon-inducible, double-stranded RNA-dependent protein kinase, PKR
- PMID: 10400814
- PMCID: PMC112801
- DOI: 10.1128/JVI.73.8.7080-7086.1999
Human immunodeficiency virus type 1 tat protein activates transcription factor NF-kappaB through the cellular interferon-inducible, double-stranded RNA-dependent protein kinase, PKR
Abstract
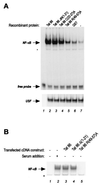
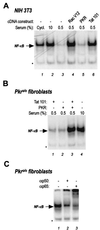
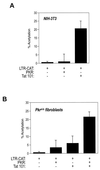
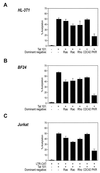
The transactivator protein of human immunodeficiency virus type 1 (HIV-1) (Tat) is a powerful activator of nuclear factor-kappaB (NF-kappaB), acting through degradation of the inhibitor IkappaB-alpha (F. Demarchi, F. d'Adda di Fagagna, A. Falaschi, and M. Giacca, J. Virol. 70:4427-4437, 1996). Here, we show that this activity of Tat requires the function of the cellular interferon-inducible protein kinase PKR. Tat-mediated NF-kappaB activation and transcriptional induction of the HIV-1 long terminal repeat were impaired in murine cells in which the PKR gene was knocked out. Both functions were restored by cotransfection of Tat with the cDNA for PKR. Expression of a dominant-negative mutant of PKR specifically reduced the levels of Tat transactivation in different human cell types. Activation of NF-kappaB by Tat required integrity of the basic domain of Tat; previous studies have indicated that this domain is necessary for specific Tat-PKR interaction.
Figures


Similar articles
-
Activation of transcription factor NF-kappaB by the Tat protein of human immunodeficiency virus type 1.J Virol. 1996 Jul;70(7):4427-37. doi: 10.1128/JVI.70.7.4427-4437.1996. J Virol. 1996. PMID: 8676466 Free PMC article.
-
Phosphorylation of HIV Tat by PKR increases interaction with TAR RNA and enhances transcription.Virol J. 2005 Feb 28;2:17. doi: 10.1186/1743-422X-2-17. Virol J. 2005. PMID: 15737233 Free PMC article.
-
RNA binding and modulation of PKR activity.Methods. 1998 Jul;15(3):189-98. doi: 10.1006/meth.1998.0623. Methods. 1998. PMID: 9735304
-
HIV-1 Tat directly interacts with the interferon-induced, double-stranded RNA-dependent kinase, PKR.Virology. 1995 Nov 10;213(2):413-24. doi: 10.1006/viro.1995.0014. Virology. 1995. PMID: 7491766
-
Regulation of the interferon-inducible eIF-2 alpha protein kinase by small RNAs.Biochimie. 1994;76(8):770-8. doi: 10.1016/0300-9084(94)90081-7. Biochimie. 1994. PMID: 7534482 Review.
Cited by
-
Enhanced NF-κB activation via HIV-1 Tat-TRAF6 cross-talk.Sci Adv. 2024 Jan 19;10(3):eadi4162. doi: 10.1126/sciadv.adi4162. Epub 2024 Jan 19. Sci Adv. 2024. PMID: 38241362 Free PMC article.
-
Impact of viral activators and epigenetic regulators on HIV-1 LTRs containing naturally occurring single nucleotide polymorphisms.Biomed Res Int. 2015;2015:320642. doi: 10.1155/2015/320642. Epub 2015 Jan 5. Biomed Res Int. 2015. PMID: 25629043 Free PMC article.
-
Reversal of diabetes through gene therapy of diabetic rats by hepatic insulin expression via lentiviral transduction.Mol Ther. 2012 May;20(5):918-26. doi: 10.1038/mt.2012.8. Epub 2012 Feb 21. Mol Ther. 2012. PMID: 22354377 Free PMC article.
-
An NF-kappa B-dependent survival pathway protects against cell death induced by TVB receptors for avian leukosis viruses.J Virol. 2002 Jun;76(11):5581-7. doi: 10.1128/jvi.76.11.5581-5587.2002. J Virol. 2002. PMID: 11991986 Free PMC article.
-
Defining the molecular mechanisms of HIV-1 Tat secretion: PtdIns(4,5)P2 at the epicenter.Traffic. 2018 Apr 30:10.1111/tra.12578. doi: 10.1111/tra.12578. Online ahead of print. Traffic. 2018. PMID: 29708629 Free PMC article. Review.
References
-
- Alcamí J, Laín de Lera T, Folgueira L, Pedraza M A, Jacqué J-M, Bachelerie F, Noriega A R, Hay R T, Harrich D, Gaynor R B, Virelizier J-L, Arenzana-Seisdos F. Absolute dependence on κB responsive elements for initiation and Tat-mediated amplification of HIV transcription in blood CD4 T lymphocytes. EMBO J. 1995;14:1552–1560. - PMC - PubMed
-
- Baeuerle P A, Baltimore D. NF-κB: ten years after. Cell. 1996;87:13–20. - PubMed
-
- Benkirane M, Chun R F, Xiao H, Ogryzko V V, Howard B H, Nakatani Y, Jeang K T. Activation of integrated provirus requires histone acetyltransferase. p300 and P/CAF are coactivators for HIV-1 Tat. J Biol Chem. 1998;273:24898–24905. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

