Mutations in the essential spindle checkpoint gene bub1 cause chromosome missegregation and fail to block apoptosis in Drosophila
- PMID: 10402457
- PMCID: PMC2199734
- DOI: 10.1083/jcb.146.1.13
Mutations in the essential spindle checkpoint gene bub1 cause chromosome missegregation and fail to block apoptosis in Drosophila
Abstract
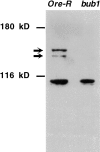
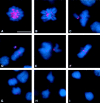
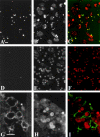
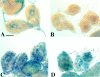
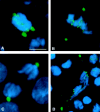
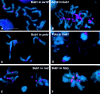
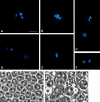
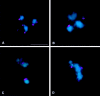
We have characterized the Drosophila mitotic checkpoint control protein Bub1 and obtained mutations in the bub1 gene. Drosophila Bub1 localizes strongly to the centromere/kinetochore of mitotic and meiotic chromosomes that have not yet reached the metaphase plate. Animals homozygous for P-element-induced, near-null mutations of bub1 die during late larval/pupal stages due to severe mitotic abnormalities indicative of a bypass of checkpoint function. These abnormalities include accelerated exit from metaphase and chromosome missegregation and fragmentation. Chromosome fragmentation possibly leads to the significantly elevated levels of apoptosis seen in mutants. We have also investigated the relationship between Bub1 and other kinetochore components. We show that Bub1 kinase activity is not required for phosphorylation of 3F3/2 epitopes at prophase/prometaphase, but is needed for 3F3/2 dephosphorylation at metaphase. Neither 3F3/2 dephosphorylation nor loss of Bub1 from the kinetochore is a prerequisite for anaphase entry. Bub1's localization to the kinetochore does not depend on the products of the genes zw10, rod, polo, or fizzy, indicating that the kinetochore is constructed from several independent subassemblies.
Figures


References
-
- Ault J.G., Lin H.P. Bivalent behaviour in Drosophila melanogaster males containing the In(1)sc4Lsc8RXchromosome. Chromosoma. 1984;90:222–228. - PubMed
-
- Ault J.G., Nicklas R.B. Tension, microtubule rearrangements, and the proper distribution of chromosomes in mitosis. Chromosoma. 1989;98:33–39. - PubMed
-
- Basu, J. 1999. The spindle assembly checkpoint in Drosophila. Ph.D. thesis. Cornell University, Ithaca, NY.
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

