Chromatin opening and transactivator potentiation by RAP1 in Saccharomyces cerevisiae
- PMID: 10409719
- PMCID: PMC84371
- DOI: 10.1128/MCB.19.8.5279
Chromatin opening and transactivator potentiation by RAP1 in Saccharomyces cerevisiae
Abstract
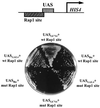
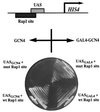
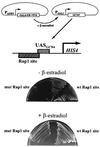
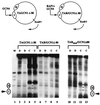
Transcriptional activators function in vivo via binding sites that may be packaged into chromatin. Here we show that whereas the transcriptional activator GAL4 is strongly able to perturb chromatin structure via a nucleosomal binding site in yeast, GCN4 does so poorly. Correspondingly, GCN4 requires assistance from an accessory protein, RAP1, for activation of the HIS4 promoter, whereas GAL4 does not. The requirement for RAP1 for GCN4-mediated HIS4 activation is dictated by the DNA-binding domain of GCN4 and not the activation domain, suggesting that RAP1 assists GCN4 in gaining access to its binding site. Consistent with this, overexpression of GCN4 partially alleviates the requirement for RAP1, whereas HIS4 activation via a weak GAL4 binding site requires RAP1. RAP1 is extremely effective at interfering with positioning of a nucleosome containing its binding site, consistent with a role in opening chromatin at the HIS4 promoter. Furthermore, increasing the spacing between binding sites for RAP1 and GCN4 by 5 or 10 bp does not impair HIS4 activation, indicating that cooperative protein-protein interactions are not involved in transcriptional facilitation by RAP1. We conclude that an important role of RAP1 is to assist activator binding by opening chromatin.
Figures



References
-
- Bortvin, A. Unpublished data.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
