The Wnt/Wg signal transducer beta-catenin controls fibronectin expression
- PMID: 10409747
- PMCID: PMC84410
- DOI: 10.1128/MCB.19.8.5576
The Wnt/Wg signal transducer beta-catenin controls fibronectin expression
Abstract
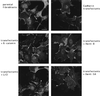
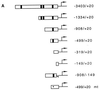
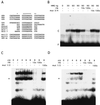
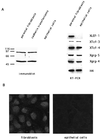
beta-Catenin stabilizes the cadherin cell adhesion complex but, as a component of the Wnt/Wg signaling pathway, also controls gene expression by forming a heterodimer with a transcription factor of the LEF-TCF family. We demonstrate that the substrate adhesion molecule fibronectin is a direct target of Wnt/Wg signaling. Nuclear depletion of beta-catenin following cadherin transfection in Xenopus fibroblasts resulted in downregulation of fibronectin expression which was restored by activating the Wnt/Wg signaling cascade via LiCl treatment or transfection of either Xwnt-8 or beta-catenin. We isolated the Xenopus fibronectin gene (FN) promoter and found four putative LEF-TCF binding sites. By comparing the activities of different fibronectin gene reporter constructs in fibroblasts and cadherin transfectants, the LEF-TCF site at position -368 was identified as a Wnt/Wg response element. LEF-1-related proteins were found in nuclei of the fibroblasts but were absent in a kidney epithelial cell line. Consistent with the lack of these transcription factors, the FN promoter was silent in the epithelial cells but was activated upon transfection of LEF-1. Wild-type Xenopus Tcf-3 (XTcf-3) was unable to activate FN promoter reporter constructs, while a mutant lacking the groucho binding region behaved like LEF-1. In contrast to XTcf-3, LEF-1 does not interact with groucho proteins, which turn TCFs into activators or repressors (J. Roose, M. Molenaar, J. Hurenkamp, J. Peterson, H. Brantjes, P. Moerer, M. van de Wetering, O. Destreé, and H. Clevers, Nature 395:608-612, 1998). Together these data provide evidence that expressing LEF-1 enables fibroblasts, in contrast to epithelial cells, to respond to the Wnt/Wg signal via beta-catenin in stimulating fibronectin gene transcription. Our findings further promote the idea that due to its dual function, beta-catenin regulates the balance between cell-cell and cell-substrate adhesion.
Figures







Similar articles
-
Functional diversity of Xenopus lymphoid enhancer factor/T-cell factor transcription factors relies on combinations of activating and repressing elements.J Biol Chem. 2002 Apr 19;277(16):14159-71. doi: 10.1074/jbc.M107055200. Epub 2002 Jan 30. J Biol Chem. 2002. PMID: 11821382
-
The C-terminal transactivation domain of beta-catenin is necessary and sufficient for signaling by the LEF-1/beta-catenin complex in Xenopus laevis.Mech Dev. 1999 Mar;81(1-2):65-74. doi: 10.1016/s0925-4773(98)00225-1. Mech Dev. 1999. PMID: 10330485
-
Direct regulation of the Xenopus engrailed-2 promoter by the Wnt signaling pathway, and a molecular screen for Wnt-responsive genes, confirm a role for Wnt signaling during neural patterning in Xenopus.Mech Dev. 1999 Sep;87(1-2):21-32. doi: 10.1016/s0925-4773(99)00136-7. Mech Dev. 1999. PMID: 10495268
-
Regulation of LEF-1/TCF transcription factors by Wnt and other signals.Curr Opin Cell Biol. 1999 Apr;11(2):233-40. doi: 10.1016/s0955-0674(99)80031-3. Curr Opin Cell Biol. 1999. PMID: 10209158 Review.
-
The Yin-Yang of TCF/beta-catenin signaling.Adv Cancer Res. 2000;77:1-24. doi: 10.1016/s0065-230x(08)60783-6. Adv Cancer Res. 2000. PMID: 10549354 Review.
Cited by
-
IGF2BP1 promotes mesenchymal cell properties and migration of tumor-derived cells by enhancing the expression of LEF1 and SNAI2 (SLUG).Nucleic Acids Res. 2013 Jul;41(13):6618-36. doi: 10.1093/nar/gkt410. Epub 2013 May 15. Nucleic Acids Res. 2013. PMID: 23677615 Free PMC article.
-
Disruption of the Dapper3 gene aggravates ureteral obstruction-mediated renal fibrosis by amplifying Wnt/β-catenin signaling.J Biol Chem. 2013 May 24;288(21):15006-14. doi: 10.1074/jbc.M113.458448. Epub 2013 Apr 11. J Biol Chem. 2013. PMID: 23580654 Free PMC article.
-
Divergent regulation of Wnt-mediated development of the dorsomedial and ventrolateral dermomyotomal lips.Histochem Cell Biol. 2012 Sep;138(3):503-14. doi: 10.1007/s00418-012-0971-y. Epub 2012 Jun 6. Histochem Cell Biol. 2012. PMID: 22669461
-
Wnt/β-catenin signaling is hyperactivated in systemic sclerosis and induces Smad-dependent fibrotic responses in mesenchymal cells.Arthritis Rheum. 2012 Aug;64(8):2734-45. doi: 10.1002/art.34424. Arthritis Rheum. 2012. PMID: 22328118 Free PMC article.
-
Novel markers for differentiation of lobular and ductal invasive breast carcinomas by laser microdissection and microarray analysis.BMC Cancer. 2007 Mar 27;7:55. doi: 10.1186/1471-2407-7-55. BMC Cancer. 2007. PMID: 17389037 Free PMC article.
References
-
- Behrens J, von der Kries J P, Kühl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Functional interaction of β-catenin with the transcription factor LEF-1. Nature. 1996;382:638–642. - PubMed
-
- Cavallo R, Rubinstein D, Peifer M. Armadillo and dTCF: a marriage made in the nucleus. Curr Opin Genet Dev. 1997;7:459–466. - PubMed
-
- Chirgwin J M, Przybyia A E, MacDonald R J, Rutter W J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979;18:5294–5299. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
