Activity of the c-myc replicator at an ectopic chromosomal location
- PMID: 10409757
- PMCID: PMC84420
- DOI: 10.1128/MCB.19.8.5685
Activity of the c-myc replicator at an ectopic chromosomal location
Erratum in
- Mol Cell Biol 1999 Dec;19(12):8694
Abstract
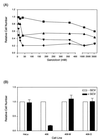
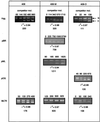
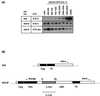
DNA replication starts at multiple discrete sites across the human chromosomal c-myc region, including two or more sites within 2.4 kb upstream of the c-myc gene. The corresponding 2.4-kb c-myc origin fragment confers autonomously replicating sequence (ARS) activity on plasmids, which specifically initiate replication in the origin fragment in vitro and in vivo. To test whether the region that displays plasmid replicator activity also acts as a chromosomal replicator, HeLa cell sublines that each contain a single copy of the Saccharomyces cerevisiae FLP recombinase target (FRT) sequence flanked by selectable markers were constructed. A clonal line containing a single unrearranged copy of the transduced c-myc origin was produced by cotransfecting a donor plasmid containing the 2.4-kb c-myc origin fragment and FRT, along with a plasmid expressing the yeast FLP recombinase, into cells containing a chromosomal FRT acceptor site. The amount of short nascent DNA strands at the chromosomal acceptor site was quantitated before and after targeted integration of the origin fragment. Competitive PCR quantitation showed that the c-myc origin construct substantially increased the amount of nascent DNA relative to that at the unoccupied acceptor site and to that after the insertion of non-myc DNA. The abundance of nascent strands was greatest close to the c-myc insert of the integrated donor plasmid, and significant increases in nascent strand abundance were observed at sites flanking the insertion. These results provide biochemical and genetic evidence for the existence of chromosomal replicators in metazoan cells and are consistent with the presence of chromosomal replicator activity in the 2.4-kb region of c-myc origin DNA.
Figures





References
-
- Aladjem M I, Rodewald L W, Kolman J L, Wahl G M. Genetic dissection of a mammalian replicator in the human beta-globin locus. Science. 1998;281:1005–1009. - PubMed
-
- Berberich S, Trivedi A, Daniel D C, Johnson E M, Leffak M. In vitro replication of plasmids containing human c-myc DNA. J Mol Biol. 1995;245:92–109. - PubMed
-
- Bielinsky A K, Gerbi S A. Discrete start sites for DNA synthesis in the yeast ARS1 origin. Science. 1998;279:95–98. - PubMed
-
- Connolly A R, Cleland L G, Kirkham B W. Mathematical considerations of competitive polymerase chain reaction. J Immunol Methods. 1995;187:201–211. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
