Nup124p is a nuclear pore factor of Schizosaccharomyces pombe that is important for nuclear import and activity of retrotransposon Tf1
- PMID: 10409764
- PMCID: PMC84427
- DOI: 10.1128/MCB.19.8.5768
Nup124p is a nuclear pore factor of Schizosaccharomyces pombe that is important for nuclear import and activity of retrotransposon Tf1
Abstract
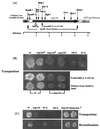
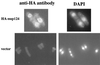
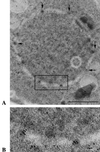
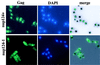
The long terminal repeat (LTR)-containing retrotransposon Tf1 propagates within the fission yeast Schizosaccharomyces pombe as the result of several mechanisms that are typical of both retrotransposons and retroviruses. To identify host factors that contribute to the transposition process, we mutagenized cultures of S. pombe and screened them for strains that were unable to support Tf1 transposition. One such strain contained a mutation in a gene we named nup124. The product of this gene contains 11 FXFG repeats and is a component of the nuclear pore complex. In addition to the reduced levels of Tf1 transposition, the nup124-1 allele caused a significant reduction in the nuclear localization of Tf1 Gag. Surprisingly, the mutation in nup124-1 did not cause any reduction in the growth rate, the nuclear localization of specific nuclear localization signal-containing proteins, or the cytoplasmic localization of poly(A) mRNA. A two-hybrid analysis and an in vitro precipitation assay both identified an interaction between Tf1 Gag and the N terminus of Nup124p. These results provide evidence for an unusual mechanism of nuclear import that relies on a direct interaction between a nuclear pore factor and Tf1 Gag.
Figures




References
-
- Aitchison J D, Blobel G, Route M P. Kap104p: a karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science. 1996;274:624–627. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
