Cell cycle regulation of Dfp1, an activator of the Hsk1 protein kinase
- PMID: 10411894
- PMCID: PMC17535
- DOI: 10.1073/pnas.96.15.8443
Cell cycle regulation of Dfp1, an activator of the Hsk1 protein kinase
Abstract
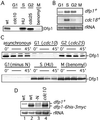
In fission yeast, the Hsk1 protein kinase is essential for the initiation of DNA replication. We have shown previously that Hsk1 forms a heterodimeric complex with the regulatory subunit, Dfp1. In this report we describe the further characterization of Dfp1. Reconstitution experiments with purified proteins indicate that Dfp1 is necessary and sufficient to activate Hsk1 phosphorylation of exogenous substrates, such as the Schizosaccharomyces pombe minichromosome maintenance protein Cdc19. The dfp1(+) gene is essential for viability of S. pombe, and depletion of the Dfp1 protein significantly delays the onset of S phase. Dfp1 is a phosphoprotein in vivo and becomes hyperphosphorylated when cells are blocked in S phase by treatment with the DNA synthesis inhibitor hydroxyurea. Hyperphosphorylation in S phase depends on the checkpoint kinase Cds1. The abundance of Dfp1 varies during progression through the cell cycle. The protein is absent when cells are arrested in G(1) phase. When cells are released into the cell cycle, Dfp1 appears suddenly at the G(1)/S transition, coincident with the initiation of DNA replication. The absence of Dfp1 before S phase is due largely, but not exclusively, to posttranscriptional regulation. We propose that cell cycle-regulated activation of Dfp1 expression at the G(1)/S transition results in activation of the Hsk1 protein kinase, which, in turn, leads to the initiation of DNA replication.
Figures

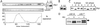

References
-
- Culotti J, Hartwell L H. Exp Cell Res. 1971;67:389–401. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

