Targeted manipulation of maize genes in vivo using chimeric RNA/DNA oligonucleotides
- PMID: 10411950
- PMCID: PMC17591
- DOI: 10.1073/pnas.96.15.8768
Targeted manipulation of maize genes in vivo using chimeric RNA/DNA oligonucleotides
Abstract
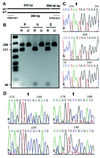
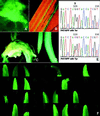
Site-specific heritable mutations in maize genes were engineered by introducing chimeric RNA/DNA oligonucleotides. Two independent targets within the endogenous maize acetohydroxyacid synthase gene sequence were modified in a site-specific fashion, thereby conferring resistance to either imidazolinone or sulfonylurea herbicides. Similarly, an engineered green fluorescence protein transgene was site-specifically modified in vivo. Expression of the introduced inactive green fluorescence protein was restored, and plants containing the modified transgene were regenerated. Progeny analysis indicated Mendelian transmission of the converted transgene. The efficiency of gene conversion mediated by chimeric oligonucleotides in maize was estimated as 10(-4), which is 1-3 orders of magnitude higher than frequencies reported for gene targeting by homologous recombination in plants. The heritable changes in maize genes engineered by this approach create opportunities for basic studies of plant gene function and agricultural trait manipulation and also provide a system for studying mismatch repair mechanisms in maize.
Figures


Comment in
-
Gene therapy in plants.Proc Natl Acad Sci U S A. 1999 Jul 20;96(15):8321-3. doi: 10.1073/pnas.96.15.8321. Proc Natl Acad Sci U S A. 1999. PMID: 10411868 Free PMC article. No abstract available.
References
-
- Ohl S, Offringa R, van den Elzen P J M, Hooykaas P J J. In: Homologous Recombination and Gene Silencing in Plants. Paszkowski J, editor. Dordrecht, The Netherlands: Kluwer; 1994. pp. 191–217.
-
- Puchta H. Trends Plant Sci. 1998;3:77–78.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

